2. Pre-processing¶
Author: Valentin ChristiaensSuitable for VIP v1.1.3 onwardsLast update: 2022/03/25
Table of contents
- 2.1. Recentering coronagraphic NACO data
- 2.2. Recentering coronagraphic SPHERE/IFS data
- 2.3. Pre-processing a SINFONI datacube
The subpackage preproc contains all sorts of low-level image transformation routines: translation, rotation, cropping, pixel resampling, etc. It also contains functions dedicated to the preparation of 3D or 4D arrays before stellar halo modeling and subtraction (i.e. post-processing). In this tutorial we’ll show how to use some of these pre-processing routines to perform the following tasks: - recentering a NACO L’ coronagraphic dataset and trimming bad frames out; - correcting bad pixels
from a SPHERE/IFS coronagraphic datacube and recentering it; - perform a “full pre-processing” of non-coronagraphic SINFONI data on HD 179218.
It is recommended to perform (low-level) calibration tasks such as dark subtraction or flat-fielding using the observatory pipeline of the respective instrument the data were acquired with, before using the pre-processing routines of VIP. The only potential exception would be for the use of a PCA-based sky-subtraction routine implemented in VIP, vip.preproc.cube_subtract_sky_pca, which is not showcased in this tutorial but typically outperforms classical median-sky subtraction at thermal
IR wavelengths (e.g. Hunziker et al. 2018).
Let’s first import a couple of external packages needed in this tutorial:
[1]:
%matplotlib inline
from hciplot import plot_frames, plot_cubes
from matplotlib.pyplot import *
from matplotlib import pyplot as plt
import numpy as np
from packaging import version
In the following box we import all the VIP routines that will be used in this tutorial:
[2]:
import vip_hci as vip
vvip = vip.__version__
print("VIP version: ", vvip)
if version.parse(vvip) < version.parse("1.1.3"):
msg = "Please upgrade your version of VIP"
msg+= "It should be 1.1.3 or above to run this notebook."
raise ValueError(msg)
else:
if version.parse(vvip) >= version.parse("1.3.1"):
from vip_hci.preproc import frame_fix_badpix_fft, cube_fix_badpix_interp
else:
from vip_hci.preproc import cube_fix_badpix_with_kernel
from vip_hci.config import VLT_NACO, VLT_SPHERE_IFS, VLT_SINFONI
from vip_hci.fm import normalize_psf, frame_inject_companion
from vip_hci.psfsub import median_sub, pca
# common to all versions:
from vip_hci.fits import open_fits, write_fits, info_fits
from vip_hci.metrics import snr
from vip_hci.preproc import (approx_stellar_position, cube_correct_nan, cube_crop_frames,
cube_detect_badfr_correlation, cube_fix_badpix_annuli,
cube_fix_badpix_clump, cube_fix_badpix_isolated,
cube_px_resampling, cube_recenter_2dfit, cube_recenter_dft_upsampling,
cube_recenter_satspots, cube_recenter_via_speckles, cube_shift,
frame_center_radon, frame_crop, frame_fix_badpix_isolated)
from vip_hci.var import create_synth_psf, fit_2dairydisk, fit_2dgaussian, frame_center
VIP version: 1.3.1
2.1. Recentering coronagraphic NACO data¶
Before using ADI-based post-processing algorithms, it is critical to have the star accurately aligned at the center of all frames of the datacube, a task that can be particularly difficult for coronagraphic observations. This crucial step can however significantly increase the S/N of a putative companion in post-processed images.
preproc includes several re-centering functions, whose use will depend on the data at hand:
cube_recenter_2dfitfor positive or negative 2D Gaussian/Moffat/Airy fits to the PSF;cube_recenter_dft_upsamplingwhich aligns the images based on maximizing the cross-correlation of the DFT (Guizar et al. 2008), and recenter the star based on a 2D fit (first function);cube_recenter_radonwhich recenters the star based on the Radon transform (Pueyo et al. 2015);cube_recenter_satspotswhich recenters the star based on the location of 4 satellite-spots - the star being at the center of the spots (standard e.g. for VLT/SPHERE data);cube_recenter_via_speckleswhich first aligns frames with respect to each other based on maximizing the cross-correlation of speckles, expressed in log scale (mostly relevant for coronagraphic data).
2.1.1. Loading the data¶
In the ‘datasets’ folder of the VIP_extras repository you can find a toy ADI (Angular Differential Imaging) cube and a NACO point spread function (PSF) to demonstrate the capabilities of VIP. This is an L’-band VLT/NACO dataset of beta Pictoris published in Absil et al. (2013) obtained using the Annular Groove Phase Mask (AGPM) Vortex coronagraph. The sequence has been heavily sub-sampled temporarily to make it
smaller. The frames were also cropped to the central 101x101 area. In case you want to plug-in your cube just change the path of the following cells.
More info on this dataset is available in Tutorial 1. Quick start. Note that here we load the not yet centered ADI cube:
[3]:
psfnaco = '../datasets/naco_betapic_psf.fits'
cubename = '../datasets/naco_betapic_cube.fits'
angname = '../datasets/naco_betapic_pa.fits'
cube_orig = open_fits(cubename)
psf = open_fits(psfnaco)
pa = open_fits(angname)
derot_off = 104.84 # NACO derotator offset for this observation (Absil et al. 2013)
TN = -0.45 # Position angle of true north for NACO at the epoch of observation (Absil et al. 2013)
angs = pa-derot_off-TN
Fits HDU-0 data successfully loaded. Data shape: (61, 101, 101)
Fits HDU-0 data successfully loaded. Data shape: (39, 39)
Fits HDU-0 data successfully loaded. Data shape: (61,)
Let’s measure the FWHM by fitting a 2D Gaussian to the core of the unsaturated non-coronagraphic PSF:
[4]:
%matplotlib inline
DF_fit = fit_2dgaussian(psf, crop=True, cropsize=9, debug=True, full_output=True)
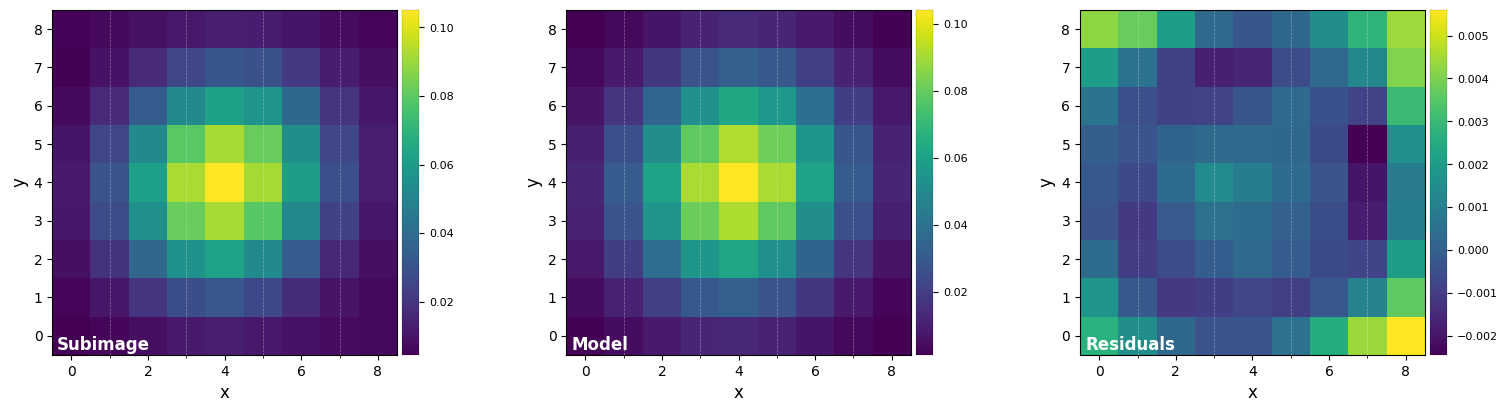
FWHM_y = 4.733218722257407
FWHM_x = 4.473682405059958
centroid y = 19.006680059041216
centroid x = 18.999424475165455
centroid y subim = 4.006680059041214
centroid x subim = 3.9994244751654535
amplitude = 0.10413004853269707
theta = -34.08563676836685
[5]:
fwhm_naco = np.mean([DF_fit['fwhm_x'],DF_fit['fwhm_y']])
print(fwhm_naco)
4.603450563658683
Let’s normalize the flux to one in a 1xFWHM aperture and crop the PSF array:
[6]:
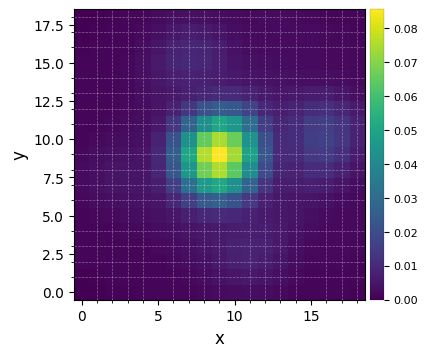
psfn = normalize_psf(psf, fwhm_naco, size=19, imlib='ndimage-fourier')
Flux in 1xFWHM aperture: 1.228
Let’s visualize the normalized PSF with hciplot.plot_frames. Feel free to adapt the backend argument throughout the notebook: 'matplotlib' allows paper-quality figures with annotations which can be saved (default), while 'bokeh' enables interactive visualization.
[7]:
plot_frames(psfn, grid=True, size_factor=4)
Let’s define the pixel scale for NACO (L’ band), which we get from a dictionary stored in the config subpackage:
[8]:
pxscale_naco = VLT_NACO['plsc']
print(pxscale_naco, "arcsec/px")
0.02719 arcsec/px
Now let’s have a look at one single image of our toy datacube. We see that the dark hole is clearly off-center:
[9]:
plot_frames(frame_crop(cube_orig[0], 15), grid=True, size_factor=4)
New shape: (15, 15)

For this NACO+AGPM dataset, three approaches could be considered for recentering - all assuming that the average location of the star throughout the observation coincides with the sweet spot (dark hole) of the coronagraph:
2.1.2. Recentering with individual 2D negative Gaussian fits¶
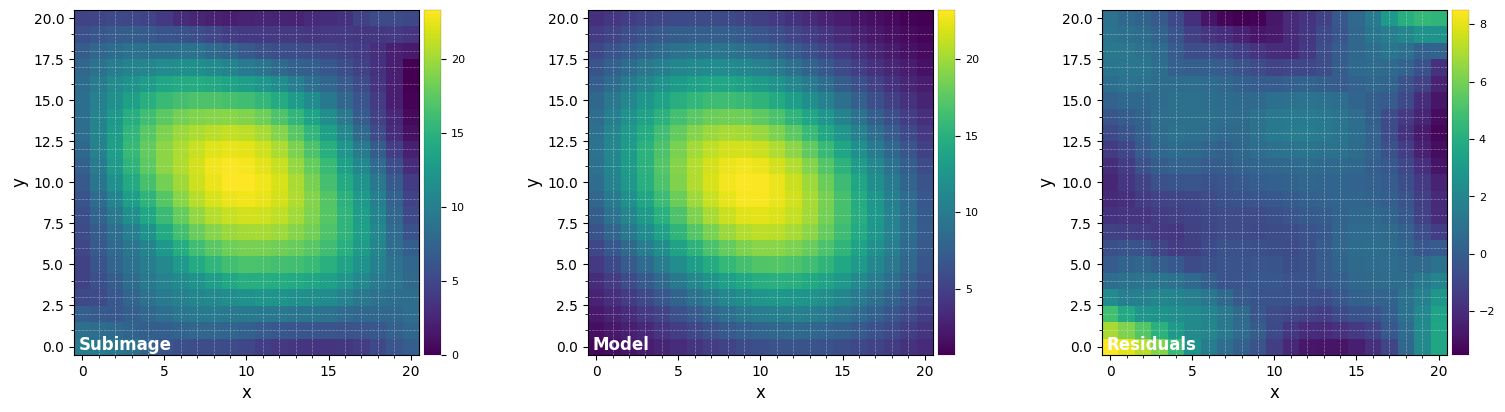
This approach consists in fitting a negative 2D Gaussian model to the dark hole area for each individual image. The coordinates of the centroid of the negative 2D Gaussian are then used to shift and align all images.
If using a different datacube, you may need to adjust the position xy of the center of the coronagraphic PSF to be fitted and the subi_size (sub-image size used for the fit, in pixels). Only use the debug mode on small datacubes (less than 20 frames), e.g. on a small subset of your datacube. In debug mode, set nproc to 1 or the plots won’t show up (values larger than 1 trigger multiprocessing).
[10]:
cube1, shy1, shx1 = cube_recenter_2dfit(cube_orig, xy=(51, 51), fwhm=fwhm_naco, nproc=1, subi_size=5,
model='gauss', negative=True, full_output=True, debug=False,
imlib='ndimage-fourier')
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:01:55
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
2d gauss-fitting
frames
0% [##############################] 100% | ETA: 00:00:00
Total time elapsed: 00:00:00
Shifting
0% [##############################] 100% | ETA: 00:00:00
Total time elapsed: 00:00:00
Running time: 0:00:00.369789
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――

2.1.3. Recentering with cross-correlation + 2D negGaussian¶
This approach is a two-step procedure involving i) a cross-correlation based method in the Fourier plane used for alignment of the images with respect to each other and ii) fitting of a negative 2D gaussian to the mean of the aligned cube for final centering.
- By not providing
subi_size(default None), the routine will not try to recenter the first frame but simply align the subsequent frames with respect to the first one. Ifsubi_sizeis an integer, it will be the size of the box used to fit a 2d gaussian to the first image, before aligning the subsequent frames to it (feel free to try running the cell below after uncommenting)
[11]:
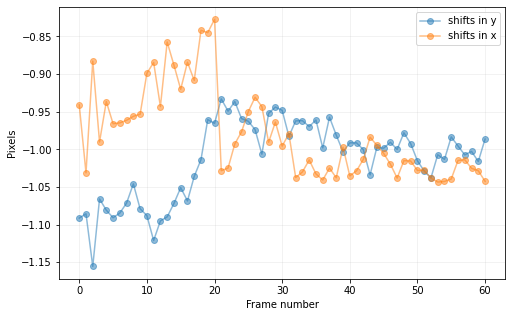
cube_align, shy2a, shx2a = cube_recenter_dft_upsampling(cube_orig, center_fr1=(51, 51),
fwhm=fwhm_naco, #subi_size=5, negative=True,
full_output=True, debug=False, imlib='ndimage-fourier')
cube_al_mean = np.mean(cube_align,axis=0)
print("Shifts required to align the frames of the cube with respect to the first one:")
plt.show()
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:01:56
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
frames
0% [##############################] 100% | ETA: 00:00:00
Total time elapsed: 00:00:00
The first frame is assumed to be well centered wrt thecenter of the array
The rest of the frames will be shifted by cross-correlation wrt the 1st
Running time: 0:00:00.505792
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Shifts required to align the frames of the cube with respect to the first one:

- we then leverage on the higher S/N in the mean image of all aligned images in order to find the final shift to place the star at the center of the images.
[12]:
_, shy2b, shx2b = cube_recenter_2dfit(cube_al_mean[np.newaxis,:,:], xy=(51, 51), fwhm=fwhm_naco,
nproc=1, subi_size=5, model='gauss', negative=True,
full_output=True, debug=False, imlib='ndimage-fourier')
print("Final shift to place the star at the center of the images:")
plt.show()
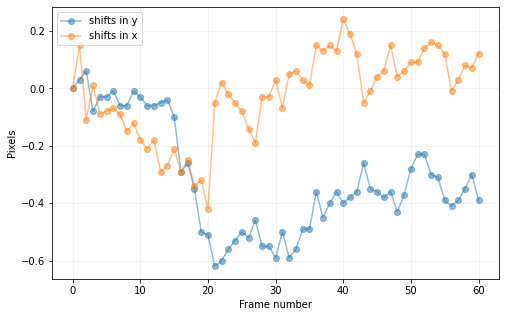
cube2 = cube_shift(cube_align, shy2b[0], shx2b[0], imlib='ndimage-fourier')
shy2 = shy2a+shy2b[0]
shx2 = shx2a+shx2b[0]
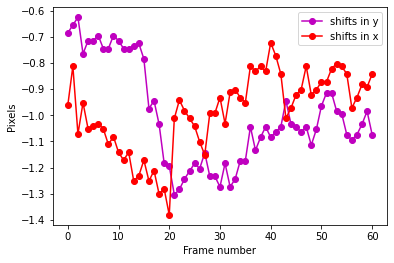
print("Combined shifts:")
plt.plot(range(cube_orig.shape[0]),shy2,'mo-', label='shifts in y')
plt.plot(range(cube_orig.shape[0]),shx2,'ro-', label='shifts in x')
plt.xlabel("Frame number")
plt.ylabel("Pixels")
plt.legend()
plt.show()
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:01:56
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
2d gauss-fitting
frames
0% [##############################] 100% | ETA: 00:00:00
Total time elapsed: 00:00:00
Shifting
0% [##############################] 100% | ETA: 00:00:00
Total time elapsed: 00:00:00
Running time: 0:00:00.036261
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Final shift to place the star at the center of the images:

Combined shifts:
2.1.4. Recentering with iterative speckle cross-correlation + 2D negGaussian¶
This approach is iterative and involves two steps at each iteration: i) the images of the cube are aligned with respect to each other based on cross-correlation of the speckles in log-scale intensity images (log scale to better capture fainter speckle with radial separation); ii) 2D negative Gaussian fit of the median image of the aligned cube. By default, it involves 5 iterations. It is well suited for coronagraphic data where an extended speckle pattern can be seen in the images in log scale and where the imprint of the coronagraph can be represented by an inverted Gaussian (e.g. NIRC2+AGPM, NACO+AGPM or ERIS+AGPM).
[13]:
cube3, _, _, shy3, shx3 = cube_recenter_via_speckles(cube_orig, alignment_iter=5, fwhm=fwhm_naco, debug=False,
recenter_median=True, fit_type='gaus', negative=True,
crop=True, subframesize=49, mask=None,
imlib='ndimage-fourier', plot=True, full_output=True)
Sub frame shape: (61, 49, 49)
Center pixel: (24, 24)
Square sum of shift vecs: 89.44960130532182
Square sum of shift vecs: 3.0493191038521013
Square sum of shift vecs: 1.0087524349940595
Square sum of shift vecs: 0.19414213562373095
Square sum of shift vecs: 0.07

2.1.5. Validation of the best recentering method¶
Frame re-centering is a case-dependent procedure. Since for this dataset there is a known companion, we can use it to infer the performance of each recentering method. All other pre-processing steps being identical, the S/N ratio of the companion is expected to be optimal with PCA-ADI (with 1 principal component used) for the cube with the best centering for the star. Not only is the companion expected to stack up in a more constructive way after derotation, but the modeling and subtraction of speckles may be slightly better if the images are perfectly aligned.
Let’s post-process each cube with PCA (npc=1), and measure the S/N ratio of Beta Pic b:
[14]:
fr1 = pca(cube1, angs)
fr2 = pca(cube2, angs)
fr3 = pca(cube3, angs)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:01:59
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
System total memory = 17.180 GB
System available memory = 0.944 GB
Done vectorizing the frames. Matrix shape: (61, 10201)
Done SVD/PCA with numpy SVD (LAPACK)
Running time: 0:00:00.128450
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Done de-rotating and combining
Running time: 0:00:01.330379
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:02:00
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
System total memory = 17.180 GB
System available memory = 0.902 GB
Done vectorizing the frames. Matrix shape: (61, 10201)
Done SVD/PCA with numpy SVD (LAPACK)
Running time: 0:00:00.115544
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Done de-rotating and combining
Running time: 0:00:01.308435
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:02:02
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
System total memory = 17.180 GB
System available memory = 0.901 GB
Done vectorizing the frames. Matrix shape: (61, 10201)
Done SVD/PCA with numpy SVD (LAPACK)
Running time: 0:00:00.116831
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Done de-rotating and combining
Running time: 0:00:01.287990
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Let’s visualize the final images:
[15]:
plot_frames((fr1, fr2, fr3), backend='bokeh')
[15]:
Let’s write down the coordinates of the point source:
[16]:
xy = (59., 35.4) # (35.8, 58.2)
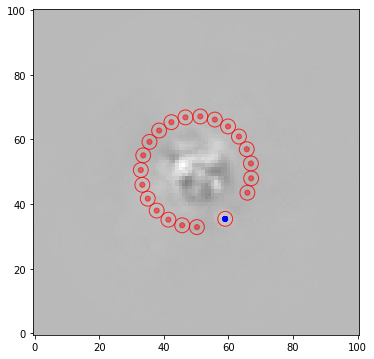
Now let’s measure the S/N ratio of the companion in each case - excluding the adjacent negative side lobes which can bias the estimate (more details in Tutorial 4. Metrics):
[17]:
snr1 = snr(fr1, xy, fwhm_naco, exclude_negative_lobes=True, plot=True)
snr2 = snr(fr2, xy, fwhm_naco, exclude_negative_lobes=True, plot=False)
snr3 = snr(fr3, xy, fwhm_naco, exclude_negative_lobes=True, plot=False)
[18]:
print(snr1, snr2, snr3)
3.095026512234823 3.540208346794688 3.6364819400156168
The third method appears to lead to a slightly better S/N ratio for Beta Pic b (all other pre- and post-processing steps being the same), although it is fair to mention that all of them yield S/N values in the same ballpark.
Based on our experience with different AGPM datasets, the iterative speckle-correlation based alignment + negative 2D Gaussian fit method is the one we recommend to use to recenter AGPM data.
[19]:
cube_cen = cube3
2.1.6. Bad frame trimming¶
ADI-based post-processing algorithms rely on angular diversity to produce a PSF model and subtract it from each image of the datacube: a putative planet rotates in the image while the stellar PSF is expected to be mostly static. The more stable the PSF in the image sequence, the better post-processing algorithms can leverage this (since it will be easier). For observations in highly stable conditions, a simple median image of the cube may be a good enough PSF model for subtraction.
Considering the above, an optimal reduction+post-processing pipeline should involve trimming bad frames. In VIP, several functions have been implemented to tackle this task:
cube_detect_badfr_ellipticityto reject bad frames based on the ellipticity of the PSF;cube_detect_badfr_pxstatsto reject bad frames based on pixel intensity statistics in a given area of the image;cube_detect_badfr_correlationto reject bad frames based on some measure of the cross-correlation or distance to a good frame.
By default, we recommend using the cross-correlation (or distance) criterion, in particular for algorithms involving a low-rank approximation of the image cube. Let’s try it out on our Beta Pic dataset, and see how trimming bad frames can affect the S/N ratio of the companion.
One can use the median image of the sequence as reference:
[20]:
good_idx_f, bad_idx_f = cube_detect_badfr_correlation(cube_cen, frame_ref=np.median(cube_cen,axis=0),
crop_size=31, dist='pearson', percentile=50,
threshold=None, plot=True, verbose=True)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:02:04
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Done detecting bad frames from cube: 31 out of 61 (50.8%)
Running time: 0:00:00.028703
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――

By default a (centered) cropped image is used for the calculation (set by crop_sz) of the Pearson correlation coefficient (PCC; dist='pearson'; although other metrics can be used). In practice, what is within 1 resolution element or hidden by a coronagraphic mask is usually not relevant, so it can be preferable to use an annular region for the calculation, set by mode='annulus', inradius and width.
The bad frame rejection can be based either on a requested percentile (e.g. percentile=30 means only the 70% most correlated frames are kept) or absolute threshold (if both are provided, threshold takes precedence).
Let’s try an absolute threshold of 0.99 in an annular region encompassing the planet:
[21]:
good_idx_a, bad_idx_a = cube_detect_badfr_correlation(cube_cen, frame_ref=np.median(cube_cen,axis=0),
crop_size=31, dist='pearson', threshold=0.99,
mode='annulus', inradius=int(fwhm_naco),
width=int(4*fwhm_naco), plot=True, verbose=True)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:02:04
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Done detecting bad frames from cube: 29 out of 61 (47.5%)
Running time: 0:00:00.032470
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――

Now let’s compare the S/N ratio of the planet obtained after PCA-ADI post-processing on: the non-trimmed cube, the cube trimmed using the PCC in the cropped sub-image, and trimmed using the PCC in an annular region:
Don’t forget to trim the associated parallactic angle vector as well:
[22]:
cube_trim_f = cube_cen[good_idx_f]
angs_trim_f = angs[good_idx_f]
cube_trim_a = cube_cen[good_idx_a]
angs_trim_a = angs[good_idx_a]
[23]:
fr_trim_f = pca(cube_trim_f, angs_trim_f)
snr_trim_f = snr(fr_trim_f, xy, fwhm_naco, exclude_negative_lobes=True, plot=False)
fr_trim_a = pca(cube_trim_a, angs_trim_a)
snr_trim_a = snr(fr_trim_a, xy, fwhm_naco, exclude_negative_lobes=True, plot=True)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:02:04
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
System total memory = 17.180 GB
System available memory = 0.884 GB
Done vectorizing the frames. Matrix shape: (30, 10201)
Done SVD/PCA with numpy SVD (LAPACK)
Running time: 0:00:00.051091
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Done de-rotating and combining
Running time: 0:00:00.668848
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:02:05
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
System total memory = 17.180 GB
System available memory = 0.884 GB
Done vectorizing the frames. Matrix shape: (32, 10201)
Done SVD/PCA with numpy SVD (LAPACK)
Running time: 0:00:00.052571
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Done de-rotating and combining
Running time: 0:00:00.662250
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――

The measured S/N ratio in each of the 3 cases is:
[24]:
print(snr3, snr_trim_f, snr_trim_a)
3.6364819400156168 3.9012923460047526 4.908796692945766
We see that appropriate bad frame trimming can indeed boost the S/N ratio of a given companion.
Feel free to test other distance metrics.
2.2. Recentering coronagraphic SPHERE/IFS data¶
2.2.1. Loading the data¶
In the ‘datasets’ folder of the VIP_extras repository you can find a SPHERE/IFS ‘CENTER’ cube obtained on PDS 70 on February 24th, 2018. This datacube contains 4 satellite spots (located along a ‘X’) which can be used to infer the location of the star behind the coronagraph (at the intersection of the diagonals). A non-coronagraphic PSF spectral cube is also available.
The point of this section is to show how to:
- correct for residual bad pixels and NaN values;
- perform a fine centering of the images based on satellite-spots (in the presence of a contaminating background star);
- perform a fine centering based on the Radon transform.
[25]:
cubename = '../datasets/sphere_ifs_PDS70_cen.fits'
psfname = '../datasets/sphere_ifs_PDS70_psf.fits'
lbdaname = '../datasets/sphere_ifs_PDS70_wl.fits'
cube = open_fits(cubename)
psfifs = open_fits(psfname)
lbda = open_fits(lbdaname)
Fits HDU-0 data successfully loaded. Data shape: (39, 199, 199)
Fits HDU-0 data successfully loaded. Data shape: (39, 31, 31)
Fits HDU-0 data successfully loaded. Data shape: (39,)
Each IFS spectral cube consists of 39 monochromatic images spread in wavelengths between the Y and J bands (‘YJ’ mode) or Y and H bands (‘YJH’ mode).
A few ‘CENTER’ cubes are usually acquired for a given object of interest, either at the very beginning and very end of the observation, or at regular intervals throughout the observation, while the rest of the spectral cubes are usually obtained without satellite spots. The star location is typically inferred in those cubes by linear interpolation of the positions derived in the ‘CENTER’ cubes.
Given the large size of typical full IFS observations, in this tutorial we only include one ‘CENTER’ cube, and hence only showcase the actual determination of stellar positions in that specific cube (i.e. no linear interpolation to infer the stellar position in the rest of the observation).
Let’s first visualize a few images from the CENTER spectral cube:
[26]:
plot_frames((cube[1], cube[19], cube[-1]))

A couple of things can be noted from the inspection of the images:
- the 4 satellite spots are located along a ‘x’ shape, and expand radially with increasing wavelength;
- a bright background star (not moving radially) is present towards the top right - its close proximity to the top-right satellite spot may interfere with an accurate retrieval of the stellar position;
- a couple of hot/cold/bad pixels remain in the image (e.g. towards the bottom left of the first frame) - these are best seen when opening the images in a FITS viewer;
- there are NaN values in the bottom right corner.
We will therefore proceed with:
- replacing NaN values;
- correcting bad pixels;
- removing a model of the background star;
- recenter the cube based on the location of the satellite spots (we will see 2 different methods).
2.2.2. Replacing NaN values¶
Some FFT-based image operations in VIP (namely sub-px shifts) do not support NaN values (interpolation-based methods are fine). Let’s replace the few NaN values at the bottom right corner of the images right away to avoid any bug during the recentering of the images.
[27]:
cube_nancorr = cube_correct_nan(cube, neighbor_box=5, min_neighbors=5)
The cube_correct_nan function replace NaN values by the median of good neighbours, iteratively until all NaN values are corrected. One can increase the box size neighbor_box (less iterations) or reduce the minimum number of good neighbours min_neighbors for a faster correction of large chunks of NaN values (consider cropping if located at the edges).
[28]:
plot_frames((cube[0], cube_nancorr[0]))

Now let’s remove the background star for a good 2D fit of all satellite spots. Note: background star overlapping with a satellite spot is an uncommon issue, feel free to skip directly to Sec. 2.2.5 if you do not have to deal with this issue, or are only interested in the recentering part.
2.2.3. Bad pixel correction¶
Several bad pixel correction algorithms are available in VIP: 1. cube_fix_badpix_isolated: to correct isolated bad pixels by sigma filtering; 2. cube_fix_badpix_annuli: which identifies bad pixels in concentric annuli (requires a circularly symmetric PSF), and replace them with the median of the value in the annulus (+ a random value following Poisson distribution); 3. cube_fix_badpix_clump: which iteratively identifies bad pixels by sigma filtering, and replace them with the median
of good neighour pixels (useful for clumps of bad pixels); 4. cube_fix_badpix_ifs: which leverages the radial expanion of the PSF with wavelength in IFS cubes to better identify bad pixels; 5. cube_fix_badpix_with_kernel: which corrects bad pixels using a 2D Gaussian kernel (requires an input bad pixel map, possibly found by one of the first 4 methods).
2.2.3.1. Sigma filter¶
Given that the bad pixels seem to be relatively isolated in the IFS images, let’s simply use the first approach (the fastest) in 2 steps. First we correct for static bad pixels, by only looking for bad pixels in the mean image of the cube (frame_by_frame=False):
[29]:
cube_sigcorr, bpm_mask = cube_fix_badpix_isolated(cube_nancorr, sigma_clip=5, num_neig=9, size=9,
frame_by_frame=False, mad=True, verbose=True,
full_output=True)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:02:07
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
processing frames
0% [##############################] 100% | ETA: 00:00:00
Total time elapsed: 00:00:01
Done replacing 2418 bad pixels using the median of neighbors
(i.e. 62 static bad pixels per channel))
Running time: 0:00:10.073418
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Next we also catch individual bad pixels (e.g. due to cosmic rays):
[30]:
cube_sigcorr, bpm_mask_tmp = cube_fix_badpix_isolated(cube_sigcorr, sigma_clip=5, num_neig=9, size=9,
frame_by_frame=True, mad=True,
verbose=True, full_output=True)
bpm_mask_2d = bpm_mask
bpm_mask = np.zeros_like(cube_nancorr)
for i in range(cube_nancorr.shape[0]):
bpm_mask[i] = bpm_mask_2d+bpm_mask_tmp[i]
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 21:02:17
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
processing frames
0% [##############################] 100% | ETA: 00:00:00
Total time elapsed: 00:02:29
Done replacing 91 bad pixels using the median of neighbors
Running time: 0:02:29.679254
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Feel free to adapt the sigma clipping threshold sigma_clip, box size for outlier identification and median filtering (through num_neig and size, respectively), and test using standard deviations instead of median absolute deviations for outlier identification (mad=False).
Let’s visualize both the static and total individual bad pixel masks (i.e. summed over all spectral channels):
[31]:
plot_frames((bpm_mask_2d, np.sum(bpm_mask_tmp,axis=0)))

Let’s now check one frame of the cube before and after bad pixel correction, and the difference between them:
[32]:
test_idx = 6
plot_frames((cube_nancorr[test_idx], cube_sigcorr[test_idx], cube_nancorr[test_idx]-cube_sigcorr[test_idx]))

Note that numba can significantly increase the speed of the bad pixel correction (numba is not a mandatory dependency of VIP, so you have to install it separately).
2.2.3.2. DFT-extrapolation¶
Another approach, provided a map of bad pixels, is to correct them with an iterative spectral deconvolution algorithm (Aach & Metzler 2001). The observed image with bad pixels can be written as g(x,y) = f(x,y) * w(x,y), where f(x,y) is the original image as it would be seen in absence of bad pixel, and w(x,y) a binary bad pixel mask. The spectral deconvolution method relies on: - the fact that the FT of a product of 2 functions is equal to the convolution of their respective FT; - the estimation of F(u,v), the FT of f(x,y), by iteratively including the most significant spectral lines from G_i(u,v), where G_i(u,v) is set to G(u,v) at the first iteration, and is then removed from its most significant mode at each subsequent iteration.
This algorithm is available in VIP (v1.3.1 onwards) in frame_fix_badpix_fft for 2D images. For 3D cubes, one can use cube_fix_badpix_interp with mode='fft'. The latter also contains the option to perform a (much faster) correction using a Gaussian kernel interpolation (illustrated in Sec. 2.3.3). Let’s consider the first 10 frames of the cube, for correction:
[33]:
if version.parse(vvip) >= version.parse("1.3.1"):
test_idx = np.arange(9)
cube_fftcorr = cube_fix_badpix_interp(cube_nancorr[test_idx], bpm_mask[test_idx], mode='fft', nit=100, nproc=None)
Let’s compare the correction obtained with the spectral deconvolution algorithm to sigma-filtering, in two regions of interest:
[34]:
if version.parse(vvip) >= version.parse("1.3.1"):
idx1 = 3
roi_y0, roi_y1 = 65, 90
roi_x0, roi_x1 = 95, 120
idx2 = 6
roi_y02, roi_y12 = 110, 135
roi_x02, roi_x12 = 60, 85
plot_frames((cube_nancorr[idx1,roi_y0:roi_y1,roi_x0:roi_x1],
cube_fftcorr[idx1,roi_y0:roi_y1,roi_x0:roi_x1],
cube_sigcorr[idx1,roi_y0:roi_y1,roi_x0:roi_x1]),
title="Region of Interest 1: no bad pix corr., DFT-based bad pix corr., sigma-filtering bad pix corr.")
plot_frames((cube_nancorr[idx2,roi_y02:roi_y12,roi_x02:roi_x12],
cube_fftcorr[idx2,roi_y02:roi_y12,roi_x02:roi_x12],
cube_sigcorr[idx2,roi_y02:roi_y12,roi_x02:roi_x12]),
title="Region of Interest 2: no bad pix corr., DFT-based bad pix corr., sigma-filtering bad pix corr.")
# difference
plot_frames((cube_fftcorr[idx1,roi_y0:roi_y1,roi_x0:roi_x1]-cube_sigcorr[idx1,roi_y0:roi_y1,roi_x0:roi_x1],
cube_fftcorr[idx2,roi_y02:roi_y12,roi_x02:roi_x12]-cube_sigcorr[idx2,roi_y02:roi_y12,roi_x02:roi_x12]),
title="Difference between DFT-based and sigma-filtering correction")
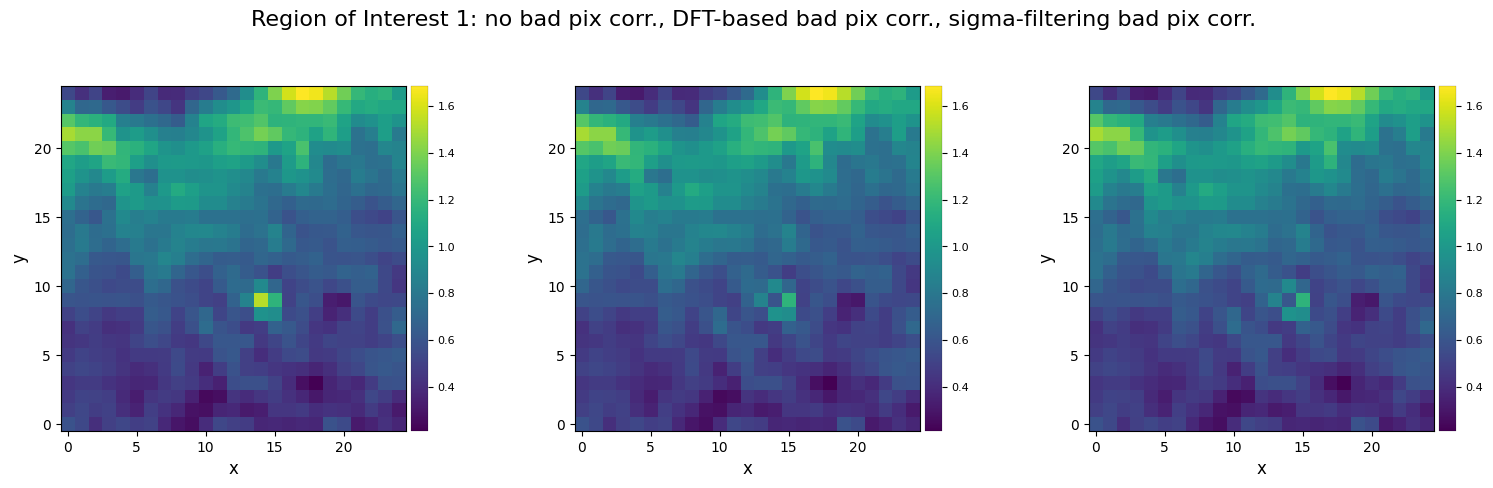


The correction is very similar in this case. Albeit slower, the spectral deconvolution algorithm can perform better for large clumps of bad pixels. Let’s illustrate that below, by artificially masking a large block of pixels, and compare the results obtained with sigma filtering and DFT-extrapolation to the original pixel intensities:
[35]:
if version.parse(vvip) >= version.parse("1.3.1"):
test_idx = 1
ty0, ty1 = 25, 29
tx0, tx1 = 159, 165
bpm_mask_test = bpm_mask.copy()
bpm_mask_test[test_idx,ty0:ty1, tx0:tx1] = 1 # set them as bad pixels even if good
[36]:
if version.parse(vvip) >= version.parse("1.3.1"):
frame_fftcorr = cube_fix_badpix_interp(cube_nancorr[test_idx], bpm_mask_test[test_idx], tol=10, nit=2000)
frame_sigcorr = frame_fix_badpix_isolated(cube_nancorr[test_idx], bpm_mask_test[test_idx],
sigma_clip=5, num_neig=9, size=9, mad=True,
verbose=True, full_output=False)
FFT-interpolation terminated after 2000 iterations (Eg=125.06167102229024)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-07 22:24:23
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Done replacing 89 bad pixels using the median of neighbors
Running time: 0:00:00.042117
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
[38]:
if version.parse(vvip) >= version.parse("1.3.1"):
roi_y0, roi_y1 = ty0 - 10, ty1 + 10
roi_x0, roi_x1 = tx0 - 10, tx1 + 9
plot_frames((cube_nancorr[test_idx,roi_y0:roi_y1,roi_x0:roi_x1],
frame_sigcorr[roi_y0:roi_y1,roi_x0:roi_x1],
frame_fftcorr[roi_y0:roi_y1,roi_x0:roi_x1]),
title="Region of Interest 3: Original, sigma-filtering clump correction, and DFT-based clump correction, ")

Note that cube_fix_badpix_ifs would have been another option to correct for bad pixels in this IFS dataset, provided that the images are first aligned with respect to each other.
2.2.4. Background-star removal¶
[39]:
cube_corr = cube_sigcorr
We’ll use the non-coronagraphic PSF observed during the same night to subtract the background star. Let’s first normalize the observed non-coronagraphic PSF to an integrated flux of 1 within 1 FWHM. By default the FWHM is found by fitting a 2D Gaussian model with the normalize_psf function:
[40]:
crop_sz = 15
norm_psf, flux, fwhm = normalize_psf(psfifs, fwhm='fit', full_output=True, size=crop_sz)
Mean FWHM per channel:
[5.2 5.227 5.23 5.245 5.287 5.327 5.357 5.378 5.398 5.418 5.481 5.552
5.599 5.627 5.657 5.689 5.73 5.787 5.828 5.85 5.868 5.857 5.804 6.105
6.362 6.23 6.225 6.255 6.3 6.349 6.39 6.423 6.444 6.491 6.539 6.592
6.659 6.727 6.787]
Flux in 1xFWHM aperture:
[ 4999.616 8605.83 10904.33 12126.272 13194.393 14388.846 15444.562
15987.688 14864.771 12628.708 12302.344 14227.798 16315.584 17630.115
18578.563 19438.338 20021.445 20474.703 20939.023 20619.506 18458.71
13514.223 7473.132 3730.324 3097.108 4777.345 7698.074 10945.942
14079.887 17093.565 20077.491 22889.499 25092.631 26571.491 27175.983
27369.823 27411.929 27244.554 26593.662]
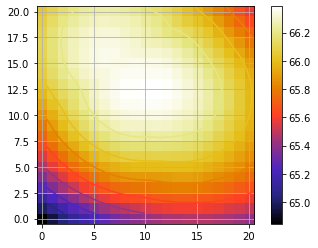
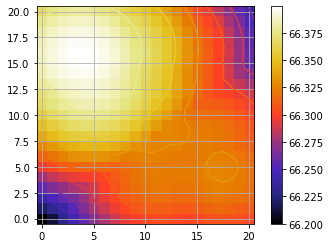
Now let’s measure the approximate position of the BKG star in the median image of the spectral cube:
[41]:
median_fr = np.nanmedian(cube_corr, axis=0)
plot_frames(median_fr, backend='bokeh')
[41]:
The approximate position is:
[42]:
bkg_xy = (144, 147)
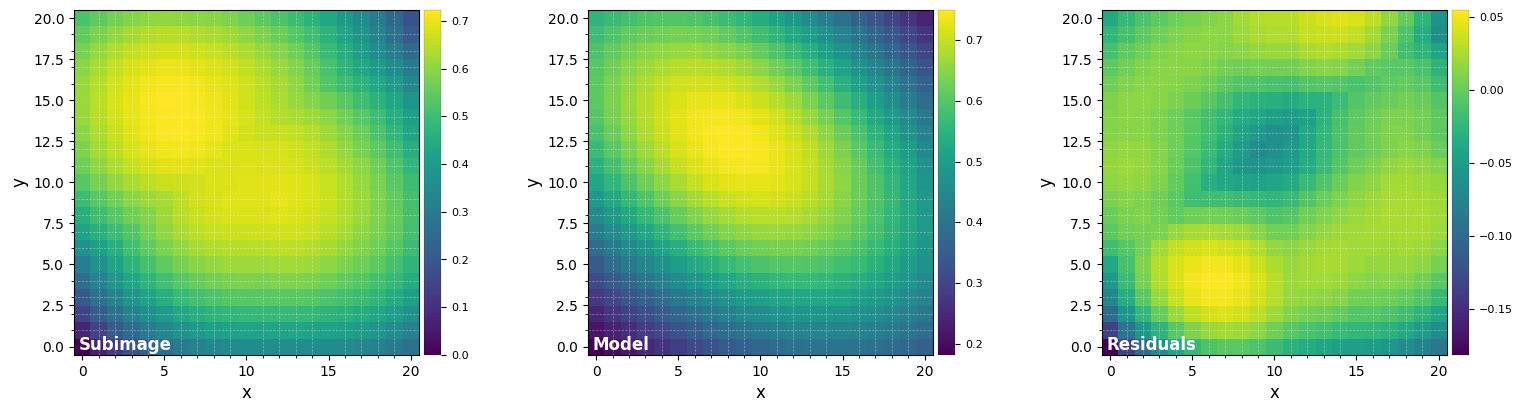
To refine the position of the background star, let’s fit a 2D Gaussian to the median frame (the debug option enables to check the residuals after model subtraction):
[43]:
med_y, med_x = fit_2dgaussian(median_fr, crop=True, cent=bkg_xy, cropsize=19, threshold=True,
sigfactor=4, full_output=False, debug=True)
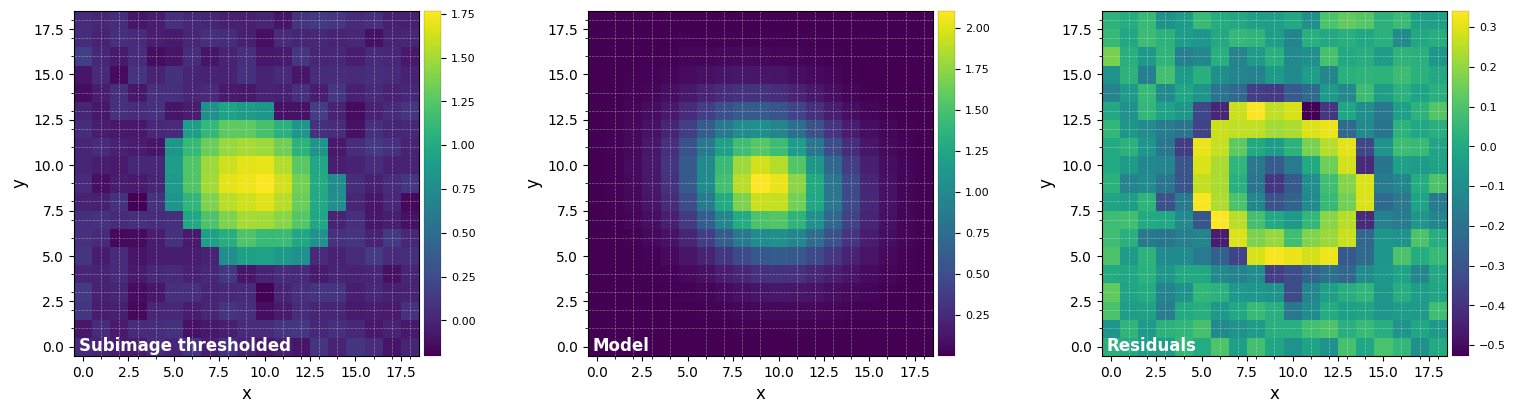
FWHM_y = 5.671741998677175
FWHM_x = 6.729458827316494
centroid y = 146.99965864506422
centroid x = 144.29842105661774
centroid y subim = 8.99965864506421
centroid x subim = 9.29842105661774
amplitude = 2.1179930039882646
theta = -213.58902731076378
Let’s also try to fit a 2D Gaussian to the individual spectral frames - now using the full_output option to retrieve all parameters of the fit:
[44]:
fit_xy = np.zeros([cube.shape[0], 2])
fit_fwhm = np.zeros([cube.shape[0], 2])
fit_amp = np.zeros([cube.shape[0]])
fit_theta = np.zeros([cube.shape[0]])
for z in range(cube.shape[0]):
if z == 0 or z == cube.shape[0]-1:
# to avoid too many plots, let's just check the residuals for the first and last spectral frame
debug = True
else:
debug = False
res = fit_2dgaussian(cube_corr[z], crop=True, cent=(med_x, med_y), cropsize=11,
threshold=False, sigfactor=0.5, full_output=True, debug=debug)
fit_xy[z,0] = res['centroid_x']
fit_xy[z,1] = res['centroid_y']
fit_fwhm[z,0] = res['fwhm_x']
fit_fwhm[z,1] = res['fwhm_y']
fit_amp[z] = res['amplitude']
fit_theta[z] = res['theta']
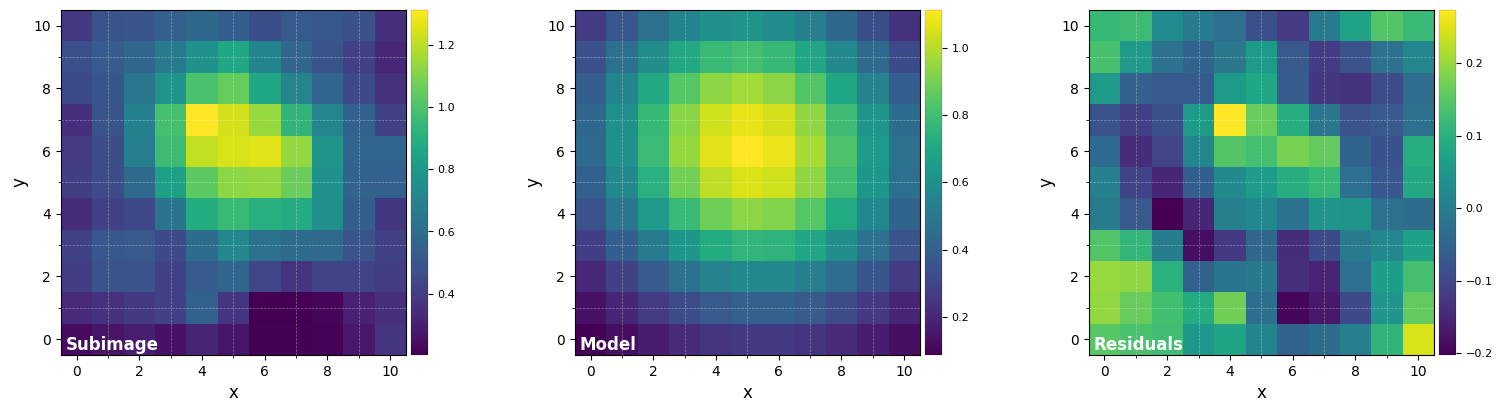
FWHM_y = 8.300835995582165
FWHM_x = 8.978869079739598
centroid y = 147.1103845672145
centroid x = 144.10591857170925
centroid y subim = 6.1103845672145045
centroid x subim = 5.105918571709249
amplitude = 1.113277303746229
theta = -209.32367802155775

FWHM_y = 10.360623229643343
FWHM_x = 9.835474386593633
centroid y = 147.28176250024754
centroid x = 144.6473909675868
centroid y subim = 6.281762500247535
centroid x subim = 5.647390967586812
amplitude = 1.8906844673410108
theta = 40.91662148035394
Let’s plot the x,y positions found by the fits:
[45]:
cols = ['b', 'r']
marks = ['o', '+']
plt.plot(range(cube.shape[0]), fit_xy[:,0], cols[0]+marks[0], label = 'x')
plt.plot(range(cube.shape[0]), fit_xy[:,1], cols[1]+marks[1], label = 'y')
plt.plot(range(cube.shape[0]), [med_x]*cube.shape[0], cols[0]+'--', label = 'med x')
plt.plot(range(cube.shape[0]), [med_y]*cube.shape[0], cols[1]+'--', label = 'med y')
plt.legend()
[45]:
<matplotlib.legend.Legend at 0x7fb49ab33df0>

Although the residual images do not show any obvious bias by the nearby satellite spot this time, the estimated position changes by up to ~1-2 px. It is unclear whether this is due to noisy data or actual jittering.
Let’s use the normalize_psf function again, this time to measure the flux in each spectral channel at the location of the BKG star. Before that we provide a sub-cube, centered at the median location of the BKG star. Internally, a step of recentering is performed (with 2D Gaussian fits similar to above) before measuring the flux in 1-FWHM radius apertures. Note that this time we fixed the value of fwhm in each spectral channels to the values found previously with the non-coronagraphic
PSF:
[46]:
subcube = cube_crop_frames(cube, size=13, xy=(med_x, med_y))
res = normalize_psf(subcube, fwhm=fwhm, threshold=None, full_output=True, verbose=True, debug=True)
fit_flux = res[1]
New shape: (39, 13, 13)
Flux in 1xFWHM aperture:
[22.308 28.005 30.229 32.877 33.838 35.297 36.041 35.814 33.682 32.372
33.429 35.369 38.005 40.719 39.875 39.969 42.366 43.676 43.503 44.432
44.149 39.168 30.354 26.05 27.228 30.263 34.66 39.877 43.732 46.068
48.592 49.766 51.57 53.283 54.694 55.647 56.211 56.871 58.021]
Let’s plot the flux inferred for the BKG star:
[47]:
cols = ['b', 'r']
marks = ['o', 'D']
plt.plot(range(cube.shape[0]), fit_flux[:], marks[0], label = 'flux')
plt.legend()
[47]:
<matplotlib.legend.Legend at 0x7fb4ba4cb970>
Now let’s use this flux to scale the normalized PSF and subtract it to each frame of the cube:
[48]:
for z in range(cube.shape[0]):
cube[z] = frame_inject_companion(cube_corr[z], norm_psf[z], med_y, med_x, -fit_flux[z],
imlib='vip-fft', interpolation='lanczos4')
Let’s check the result of the subtraction in a few frames:
[49]:
plot_frames((cube[0], cube[13], cube[25], cube[-1]))#, backend='bokeh')

Although the subtraction is not perfect (the Airy ring is not well fitted), it should be enough for the purpose of using the satellite-spots for recentering.
2.2.5. Satellite-spot based recentering¶
[50]:
xy = ((61,126), (127,138), (73,60), (140,72))
Recentering based on satellite spots is as simple as calling the cube_recenter_satspots function, providing the approximate position xy of the satellite spots in the first spectral channel and the wavelength vector lbda. Depending on your images, you may have to play with the size of the subimages subi_size where the 2D fit is peformed, the thresholding (sigfactor) or adapt the fit_type (‘moff’ or ‘gaus’). Set debug to True to actually see each 2D Moffat or Gaussian
fit (warning: in this case 4x39 Gaussian fits are performed and shown!).
[51]:
res = cube_recenter_satspots(cube, xy, lbda=lbda,
subi_size=9, sigfactor=0., fit_type='moff',
plot=True, border_mode='constant',
debug=False, verbose=False, full_output=True)
cube_sats, shift_y, shift_x, sat_y, sat_x= res

There appears to be a few outliers. Let’s confirm this with the inspection of the central area of each frame:
[52]:
from vip_hci.preproc import cube_crop_frames
plot_cubes(cube_crop_frames(cube, 141), backend='bokeh')
New shape: (39, 141, 141)
:Dataset [x,y,time] (flux)
:Cube_shape [141, 141, 39]
[52]:
Inspection of the corresponding images shows that the flux of the satellite spots is almost null in these channels (in the atmosphere’s water absorption band). One can either make the assumption that the shift is the same at all wavelengths, or interpolate from the values found in bright spectral channels.
[53]:
med_sh_y = np.median(shift_y)
med_sh_x = np.median(shift_x)
print("Median shifts (xy):", med_sh_x, med_sh_y)
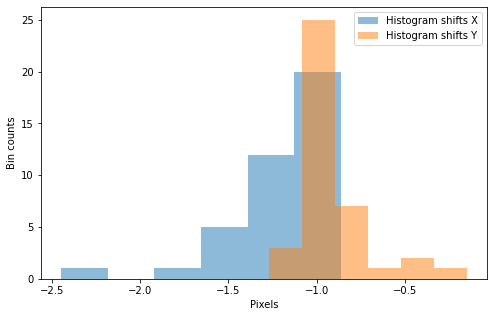
Median shifts (xy): -1.1002980518240406 -1.0058215324109199
Note that the same function can be used for recentering of IRDIS images using satellite spots.
2.2.6. Radon-transform based recentering¶
This method relies on the Radon transform, the relevant image transformation to exploit the radial information present in broad-band (or spectrally averaged) images, in order to find the location of the star. More details are available in Pueyo et al. (2015).
[54]:
mean_fr = np.mean(cube[:30], axis=0) #we skip the last channels where the BKG PSF was the most poorly subtracted
To improve the performance of the Radon-transform based algorithm, it is important to high-pass filter the input frame:
[55]:
mean_fr_hpf = vip.var.frame_filter_highpass(mean_fr, mode='gauss-subt', fwhm_size = 2*np.mean(fwhm))
[56]:
plot_frames((mean_fr,mean_fr_hpf))#, backend='bokeh')

[57]:
opt_y, opt_x = frame_center_radon(mean_fr_hpf, cropsize=131, hsize_ini=1.5, step_ini=0.1, mask_center=40,
nproc=None, satspots_cfg=None, full_output=False, verbose=True, n_iter=5,
tol=0.05, plot=True, debug=True)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-08 00:27:26
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
*** Iteration 1/5 ***
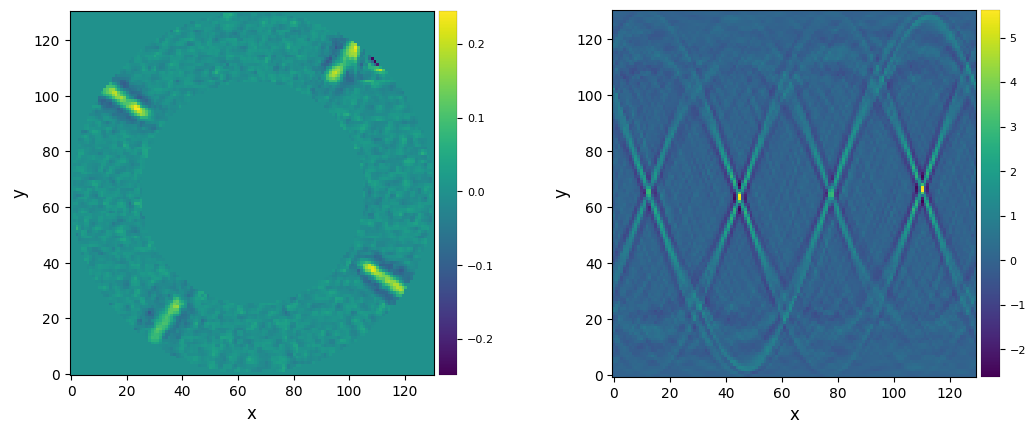
54.749336
Done 961 radon transform calls distributed in 5 processes

FWHM_y = 30.464377230585136
FWHM_x = 33.70812174915278
centroid y = 3.6811066450212437
centroid x = 5.613430474106611
centroid y subim = 3.6811066450212437
centroid x subim = 5.613430474106611
amplitude = 22.382062083689203
theta = 50.2659787023852
Cost function max: 63.11419841920198
Finished grid search radon optimization. dy=-1.132, dx=-0.939
Running time: 0:00:12.927229
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
*** Iteration 2/5 ***

64.43562202597907
Done 441 radon transform calls distributed in 5 processes
FWHM_y = 17.54288209238237
FWHM_x = 12.956221143463365
centroid y = 9.77399953901448
centroid x = 9.387563050249463
centroid y subim = 9.77399953901448
centroid x subim = 9.387563050249463
amplitude = 23.259004500533003
theta = -2283.3735778822365
Cost function max: 66.26863240417632
Finished grid search radon optimization. dy=-0.051, dx=-0.139
Running time: 0:00:19.538710
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
*** Iteration 3/5 ***
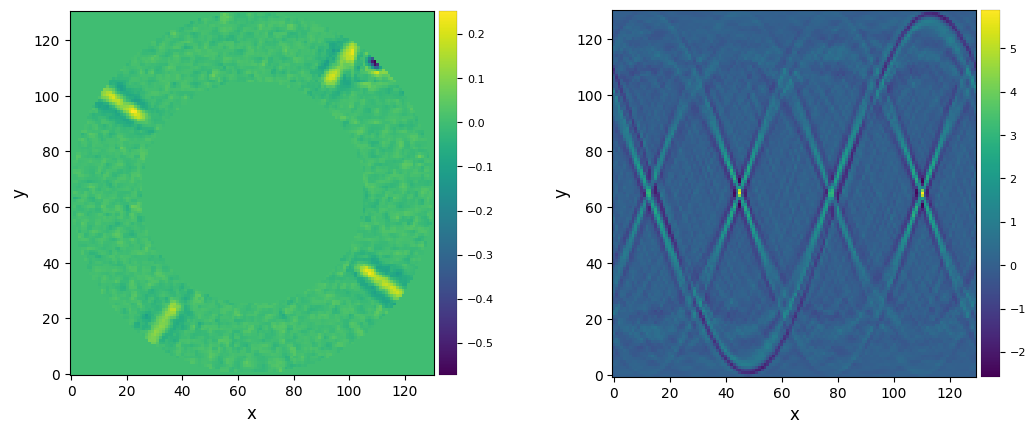
64.3905143099982
Done 441 radon transform calls distributed in 5 processes

FWHM_y = 33.152529622991764
FWHM_x = 21.784000890105098
centroid y = 13.714651346359988
centroid x = 9.071894880678963
centroid y subim = 13.714651346359988
centroid x subim = 9.071894880678963
amplitude = 1.6080336370838786
theta = -117.57484005945062
Cost function max: 66.38626859790094
Finished grid search radon optimization. dy=0.103, dx=-0.026
Running time: 0:00:26.170725
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
*** Iteration 4/5 ***

64.46047571241445
Done 441 radon transform calls distributed in 5 processes

FWHM_y = 20.901873467608077
FWHM_x = 33.87444570708223
centroid y = 12.475595627621239
centroid x = 8.27083615611571
centroid y subim = 12.475595627621239
centroid x subim = 8.27083615611571
amplitude = 0.7508091326721184
theta = -576.6257072616263
Cost function max: 66.38298977155272
Finished grid search radon optimization. dy=0.051, dx=-0.036
Running time: 0:00:32.954489
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
*** Iteration 5/5 ***
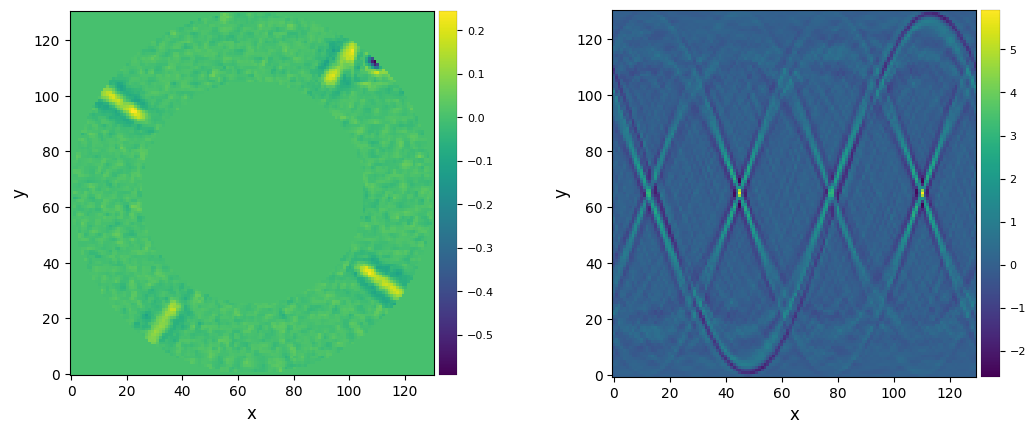
64.47481947544878
Done 441 radon transform calls distributed in 5 processes
FWHM_y = 21.867080223723267
FWHM_x = 59.346490843440705
centroid y = 22.485308859868105
centroid x = -3.2147876764907894
centroid y subim = 22.485308859868105
centroid x subim = -3.2147876764907894
amplitude = 0.21033773611154805
theta = -42.53083472433958
Cost function max: 66.3992805293159
Finished grid search radon optimization. dy=0.127, dx=-0.135
Running time: 0:00:39.641281
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Star (x,y) location: 100.27, 99.90
Final cumulative (x,y) shifts: -1.27, -0.90
Note that one can increase the precision of the algorithm, by decreasing the step used in the grid.
[58]:
cy, cx = frame_center(mean_fr)
print("Estimated star coordinates (xy):", opt_x, opt_y)
print("Estimated shifts (xy):", cx-opt_x, cy-opt_y)
Estimated star coordinates (xy): 100.27344084236717 99.90170623308153
Estimated shifts (xy): -1.2734408423671653 -0.9017062330815264
As expected, the Radon-transform based algorithm found similar shift values as the satellite-spot based one, since the most prominent features driving the centering are the wavelength-smeared satellite spots.
Once the stellar centroid found, it is only a matter of shifting each frame of the cube with the same xy shifts:
[59]:
cy, cx = frame_center(mean_fr)
cube = cube_shift(cube_corr, cy-opt_y, cx-opt_x)
[60]:
plot_frames((cube[0], cube[15], cube[-1]))#, backend='bokeh')

2.3. Pre-processing a SINFONI datacube¶
2.3.1. Loading the data¶
In the ‘datasets’ folder of the VIP_extras repository you can find a SINFONI datacube obtained on HD 179218 in May 2014. The star did not saturate and no coronagraph was used.
The point of this section is to show how to:
- correct for residual bad pixels in images where the vertical sampling is half of the horizontal sampling;
- resample images vertically;
- perform a fine recentering of the images based on 2D Gaussian fits.
[61]:
cubename = '../datasets/sinfoni_HD179218_cube.fits'
lbdaname = '../datasets/sinfoni_HD179218_lbda.fits'
cube = open_fits(cubename)
lbda = open_fits(lbdaname)
Fits HDU-0 data successfully loaded. Data shape: (400, 64, 64)
Fits HDU-0 data successfully loaded. Data shape: (400,)
Each original spectral cube consisted of ~2000 monochromatic images spread in wavelengths over the H and K bands.
Given the large size of typical full SINFONI observations, in this tutorial we only include one datacube, obtained after calibration using the ESO pipeline (including dark subtraction, flat-fielding, sky subtraction, wavelength calibration and 3D spectral cube synthesis), and after cropping the cube to only keep the last 400 channels. Note that the procedure below would be identical with a non-cropped cube.
Let’s first visualize a few images from the datacube:
[62]:
plot_frames((cube[1], cube[200], cube[-1]))
A couple of things can be noted from the inspection of the images:
- the PSF is not centered and expands radially with increasing wavelength;
- the vertical sampling of pixels is half that of the horizontal sampling (i.e. every pair of rows is identical);
- there are bad pixels in the images (best seen with harsher cuts) which tend to come as clumps or horizontal streaks.
We will therefore proceed with:
- Measuring the FWHM of the PSF in each frame;
- identifying and correcting bad pixels;
- resample the images vertically;
- recenter the cube based on 2D Gaussian fits.
2.3.2. FWHM measurement¶
As seen previously, the FWHM can be easily measured in each frame using the normalize_psf function. Let’s first consider a 2D Gaussian fit to the mean image, in order to find the average position of the star:
[63]:
mean_fr = np.mean(cube,axis=0)
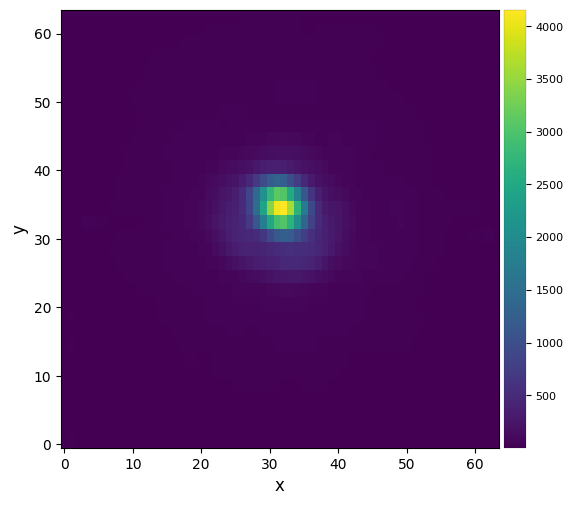
plot_frames(mean_fr)#, backend='bokeh')
[64]:
mean_y, mean_x = fit_2dgaussian(mean_fr, crop=True, cent=(33,32), cropsize=20, threshold=True,
sigfactor=5, full_output=False, debug=True)
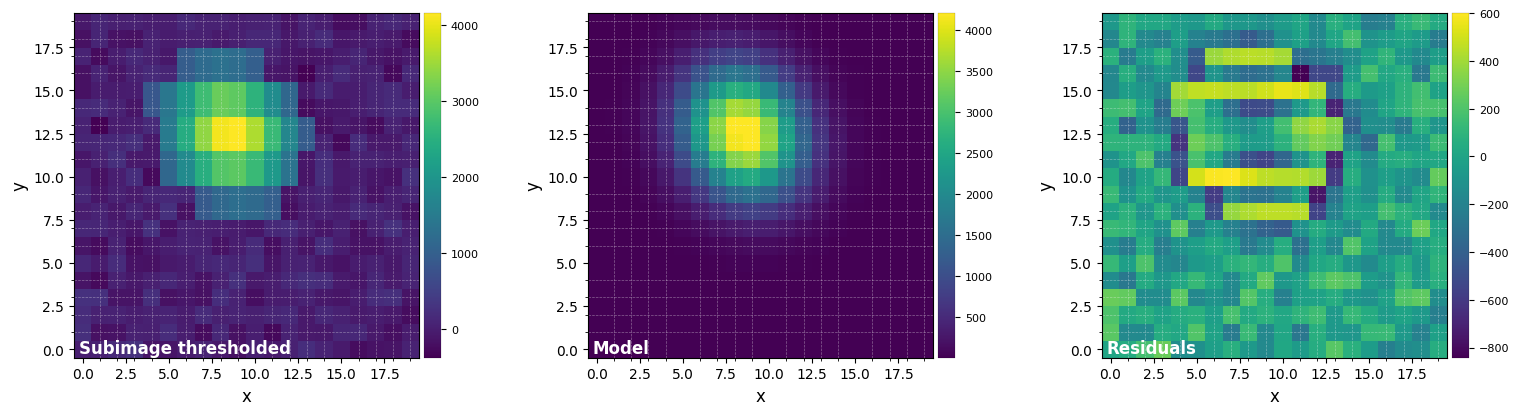
FWHM_y = 5.1970142632260625
FWHM_x = 5.9996954004492515
centroid y = 34.54708052576598
centroid x = 31.531041790076145
centroid y subim = 12.547080525765978
centroid x subim = 8.531041790076145
amplitude = 4376.261962567475
theta = 118.4945957768324
Now let’s consider a sub-cube centered on the mean position, and use normalize_psf to measure FWHM and flux:
[65]:
subcube = cube_crop_frames(cube, size=21, xy=(mean_x, mean_y), force=True) # force cropping to odd size from even size
res = normalize_psf(subcube, fwhm='fit', full_output=True)
norm_psf, fluxes, fwhm = res
WARNING: `size` is odd while input frame size is even. Make sure the center coordinates are set properly
New shape: (400, 21, 21)
Mean FWHM per channel:
[6.585 6.606 6.59 6.593 6.579 6.58 6.594 6.583 6.598 6.591 6.593 6.593
6.593 6.602 6.588 6.6 6.595 6.61 6.593 6.593 6.602 6.584 6.595 6.606
6.603 6.598 6.593 6.613 6.593 6.604 6.613 6.61 6.608 6.595 6.609 6.608
6.594 6.616 6.608 6.625 6.592 6.619 6.625 6.604 6.629 6.605 6.615 6.606
6.618 6.633 6.602 6.637 6.62 6.636 6.619 6.629 6.619 6.613 6.633 6.615
6.624 6.634 6.62 6.63 6.618 6.626 6.628 6.621 6.629 6.62 6.631 6.635
6.627 6.634 6.636 6.629 6.64 6.646 6.646 6.629 6.631 6.633 6.639 6.647
6.633 6.644 6.645 6.64 6.637 6.627 6.637 6.639 6.638 6.656 6.633 6.663
6.651 6.641 6.652 6.637 6.656 6.647 6.66 6.64 6.633 6.659 6.667 6.66
6.654 6.65 6.66 6.652 6.663 6.652 6.655 6.664 6.666 6.664 6.649 6.659
6.667 6.657 6.662 6.667 6.668 6.675 6.678 6.669 6.668 6.657 6.661 6.666
6.675 6.663 6.677 6.671 6.68 6.661 6.665 6.678 6.676 6.675 6.676 6.683
6.669 6.697 6.686 6.687 6.677 6.675 6.68 6.691 6.689 6.698 6.678 6.702
6.679 6.706 6.676 6.696 6.697 6.681 6.694 6.685 6.695 6.693 6.691 6.704
6.678 6.714 6.695 6.691 6.689 6.698 6.708 6.698 6.711 6.707 6.71 6.714
6.706 6.706 6.71 6.697 6.709 6.701 6.699 6.715 6.697 6.737 6.694 6.701
6.724 6.715 6.716 6.725 6.688 6.714 6.661 6.73 6.714 6.711 6.71 6.709
6.729 6.744 6.711 6.717 6.733 6.716 6.734 6.721 6.721 6.715 6.732 6.743
6.735 6.723 6.733 6.724 6.701 6.758 6.717 6.737 6.753 6.706 6.746 6.735
6.698 6.803 6.714 6.761 6.72 6.745 6.729 6.753 6.707 6.755 6.727 6.717
6.746 6.752 6.723 6.736 6.787 6.737 6.753 6.755 6.754 6.73 6.753 6.747
6.749 6.769 6.747 6.79 6.723 6.731 6.77 6.759 6.754 6.765 6.774 6.786
6.718 6.763 6.797 6.733 6.778 6.775 6.736 6.82 6.743 6.76 6.798 6.736
6.791 6.766 6.779 6.781 6.739 6.831 6.771 6.759 6.785 6.785 6.776 6.79
6.787 6.752 6.792 6.766 6.788 6.788 6.765 6.843 6.776 6.784 6.794 6.76
6.82 6.806 6.775 6.773 6.788 6.801 6.816 6.755 6.82 6.821 6.784 6.825
6.798 6.807 6.801 6.753 6.866 6.79 6.739 6.872 6.889 6.766 6.81 6.769
6.817 6.849 6.861 6.764 6.824 6.813 6.816 6.828 6.808 6.806 6.827 6.851
6.773 6.836 6.803 6.848 6.788 6.829 6.803 6.815 6.807 6.812 6.824 6.813
6.82 6.808 6.799 6.823 6.803 6.791 6.881 6.813 6.812 6.82 6.95 6.794
6.947 6.826 6.843 6.822 6.834 6.837 6.823 6.847 6.826 6.848 6.851 6.82
6.871 6.834 6.825 6.843 6.82 6.825 6.936 6.789 6.91 6.846 6.846 6.872
6.868 6.832 6.868 6.789 6.965 6.744 6.968 6.801 6.907 6.83 6.886 6.872
6.839 6.944 6.804 6.875]
Flux in 1xFWHM aperture:
[ 97417.897 96158.095 99477.392 99980.111 101012.75 102160.009
102327.941 100876.521 99430.314 96506.986 94799.241 93850.152
97207.058 100849.865 98795.688 98721.071 97208.779 97217.461
101196.746 103418.775 103840.653 102992.356 101202.656 97581.807
92367.592 94260.867 100415.719 99966.605 98775.974 102310.906
101998.077 99730.954 97060.392 100745.204 102142.219 103102.846
102442.581 98402.309 91359.16 91201.706 96375.254 96832.811
100313.118 102635.485 103231.796 104882.547 105326.438 104920.783
98736.602 98088.361 102191.852 103086.395 97680.572 91897.172
96878.086 95341.515 100560.258 100014.97 99115.301 103641.625
103711.981 104203.675 104147.908 103728.275 104436.253 102236.566
94421.961 91719.383 97760.464 100412.619 101628.323 99694.183
97432.835 101488.487 101099.085 99479.393 99139.705 95871.373
94062.676 94399.372 98718.89 102718.613 104155.191 99507.662
93557.162 95277.736 97254.074 95598.296 93553.365 97247.918
101736.758 101622.904 100506.192 100821.195 96896.727 94903.447
95606.472 98263.131 100240.81 101014.014 101388.338 101893.501
98149.144 101135.615 104419.411 104376.609 104841.085 105862.426
107907.032 108893.375 108694.517 106751.225 107504.813 109169.877
109357.691 109897.394 109674.736 109003.535 105532.299 97679.234
85888.699 76310.896 78270.015 82370.929 87624.814 94115.577
96810.095 98639.51 100021.777 100855.253 100504.783 97860.05
98748.282 99215.646 101878.621 103881.804 106781.109 105211.607
102126.329 104526.043 104670.278 106692.146 107760.596 106005.501
99704.321 100661.48 104262.413 107438.156 106762.811 104832.655
101821.249 100755.469 103292.871 105832.772 106534.996 103929.763
102990.063 101914.984 104063.538 102715.079 103987.234 106229.626
106303.021 102331.211 98073.231 99960.561 98024.675 90807.843
91540.793 94255.576 94941.465 97772.428 101032.616 102042.715
100219.165 101601.176 101133.523 96851.856 97860.524 100299.928
99236.717 97628.804 96810.754 91482.012 94133.875 99141.784
96710.69 95373.775 88731.87 89171.139 92355.731 88479.738
88306.563 93345.438 93521.441 101187.472 103116.593 102731.679
97947.975 89757.635 97601.161 101308.976 103117.088 100623.648
87307.138 90188.782 103981.12 108758.189 110326.357 109438.789
102458.736 103282.643 108733.727 109565.384 106378.943 100739.387
102103.435 106535.677 109169.919 106824.07 104940.7 101669.853
91610.499 97630.005 101650.29 97151.449 100147.756 87840.415
66373.687 62903.064 64673.404 76229.958 85651.413 92435.058
99257.409 102319.551 105067.554 103477.061 104720.261 102571.813
91336.189 89870.421 96848.324 89078.236 90749.99 103656.785
102945.205 98974.905 98593.54 102996.549 108447.582 109447.756
106247.445 94728.795 83902.573 80876.918 88499.651 101815.015
101804.241 91198.008 92197.425 99850.369 98738.128 91853.919
94705.194 107031.92 106367.272 101418.084 104243.377 95113.548
81249.153 94560.818 105472.574 105899.656 106583.071 105859.974
102865.052 104286.165 105322.022 105003.354 92051.343 86712.233
97475.221 104267.528 108004.833 108713.483 108814.154 104123.874
99367.297 106008.402 109659.764 108331.368 110157.089 110075.696
106002.77 91309.432 84588.118 96384.766 103682.273 100893.157
98770.173 87681.019 82355.729 94002.389 104330.594 107768.013
98831.584 89072.45 99753.265 107801.567 102073.011 98041.411
100993.951 104788.345 106169.699 92110.763 88068.388 98884.006
87846.975 60805.621 73864.197 96663.378 102240.994 96872.054
75045.567 55784.335 60956.797 86346.117 103036.418 108567.935
109520.92 100744.522 105536.076 110327.21 105023.584 89664.47
93173.559 102495.917 104583.217 98639.336 90207.113 100873.916
108187.618 109986.445 112633.726 113177.354 112222.041 112510.757
113253.982 114073.541 113625.297 113222.167 110703.89 97330.115
89504.233 97472.287 88790.303 64394.302 41866.742 40567.02
47063.811 76103.578 98776.773 106279.382 111805.956 113183.213
114157.425 113938.609 113775.516 114444.119 113757.766 113610.221
107169.654 98829.535 103501.409 109303.645 108861.805 105943.149
90518.057 59325.443 60701.638 71380.981 67959.607 78428.016
90972.341 103561.296 101448.255 78061.108 63999.348 54926.283
49064.648 73287.361 95125.99 102741.183 95658.549 86558.311
75373.732 78134.243 99614.8 108070.019]
2.3.3. Bad pixel correction¶
Several bad pixel correction algorithms are available in VIP:
cube_fix_badpix_isolated: to correct isolated bad pixels by sigma filtering;cube_fix_badpix_annuli: which identifies bad pixels in concentric annuli (requires a circularly symmetric PSF), and replace them with the median of the value in the annulus (+ a random value following Poisson distribution);cube_fix_badpix_clump: which iteratively identifies bad pixels by sigma filtering, and replace them with the median of good neighour pixels (useful for clumps of bad pixels);cube_fix_badpix_ifs: which leverages the radial expanion of the PSF with wavelength in IFS cubes to better identify bad pixels;cube_fix_badpix_with_kernel: which corrects bad pixels using a 2D Gaussian kernel (requires an input bad pixel map, possibly found by one of the first 4 methods).
Since bad pixels can show up as clumps or streaks in SINFONI cubes, it is recommended to use either cube_fix_badpix_annuli or cube_fix_badpix_clump to identify them iteratively.
In the example below, we will consider cube_fix_badpix_annuli in the example below, and then cube_fix_badpix_with_kernel to correct the identified bad pixels using a Gaussian kernel.
If the PSF core spreads over at least a few pixels, there is low risk for the algorithm to erroneously consider a PSF core pixel as bad. In noisy channels, the risk may be non-negligeable though. To prevent this risk, one can use the protect_mask parameter (in which case, the coordinates of the star must also be provided through the cy and cx parameters). Let’s find the approximate stellar position in each frame with the aptly-named approx_stellar_position function:
[66]:
cyx = approx_stellar_position(cube, fwhm)
cy = cyx[:,0]
cx = cyx[:,1]
Let’s now set the mask size to the median value of the FWHM:
[67]:
protect_mask = np.median(fwhm)
Given the peculiar half resolution along the vertical axis of SINFONI cubes, we will also use the half_res_y option, which in the case cube_fix_badpix_annuli will make it consider elliptical annuli in squashed images (by a factor 2 vertically) - instead of circular annuli.
A minimum threshold min_thr can be set to avoid correcting pure noise (which can happen in high-absorption atmospheric bands). Considering the typical pixel intensities in SINFONI cubes and the fact that they are already sky-subtracted images of our SINFONI cube, we opt for a value of -5.
[68]:
cube_corr, bpm_mask, annuli = cube_fix_badpix_annuli(cube, fwhm, cy=cy, cx=cx, sig=5.,
protect_mask=protect_mask, r_in_std=10, r_out_std=None,
verbose=True, half_res_y=True, min_thr=-5,
full_output=True)
************Frame # 0 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 1 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 2 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 3 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 4 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 5 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 6 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 7 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 8 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 9 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 10 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 11 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 12 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 13 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 14 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 15 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 16 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 17 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 18 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 19 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 20 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 21 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 22 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 23 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 24 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 25 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 26 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 27 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 28 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 29 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 30 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 31 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 32 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 33 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 34 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 35 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 36 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 37 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 38 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 39 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 40 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 41 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 42 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 43 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 44 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 45 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 46 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 47 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 48 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 49 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 50 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 51 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 52 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 53 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 54 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 55 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 56 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 57 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 58 *************
centroid assumed at coords: 31.0 35.0
9.0 bpix in total, and 9.0 corrected.
************Frame # 59 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 60 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 61 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 62 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 63 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 64 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 65 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 66 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 67 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 68 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 69 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 70 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 71 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 72 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 73 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 74 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 75 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 76 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 77 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 78 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 79 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 80 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 81 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 82 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 83 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 84 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 85 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 86 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 87 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 88 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 89 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 90 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 91 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 92 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 93 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 94 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 95 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 96 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 97 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 98 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 99 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 100 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 101 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 102 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 103 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 104 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 105 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 106 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 107 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 108 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 109 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 110 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 111 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 112 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 113 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 114 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 115 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 116 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 3.0 corrected.
************Frame # 117 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 118 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 119 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 120 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 121 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 122 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 123 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 124 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 125 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 126 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 127 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 128 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 129 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 130 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 131 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 132 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 133 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 134 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 135 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 136 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 137 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 138 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 139 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 140 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 141 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 142 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 143 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 144 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 145 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 146 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 147 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 148 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 149 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 150 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 151 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 152 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 153 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 154 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 155 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 156 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 157 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 158 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 159 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 160 *************
centroid assumed at coords: 31.0 35.0
0.0 bpix in total, and 0.0 corrected.
************Frame # 161 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 162 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 163 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 164 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 165 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 166 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 167 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 168 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 169 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 170 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 171 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 172 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 173 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 174 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 175 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 176 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 177 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 178 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 179 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 180 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 181 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 182 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 183 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 184 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 185 *************
centroid assumed at coords: 31.0 35.0
0.0 bpix in total, and 0.0 corrected.
************Frame # 186 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 187 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 188 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 189 *************
centroid assumed at coords: 31.0 35.0
9.0 bpix in total, and 9.0 corrected.
************Frame # 190 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 191 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 192 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 193 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 194 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 195 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 196 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 197 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 198 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 199 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 200 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 201 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 202 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 203 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 204 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 205 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 206 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 207 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 208 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 209 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 210 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 211 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 212 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 213 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 214 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 215 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 216 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 217 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 218 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 219 *************
centroid assumed at coords: 31.0 35.0
0.0 bpix in total, and 0.0 corrected.
************Frame # 220 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 221 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 222 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 223 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 224 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 225 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 226 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 227 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 228 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 229 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 230 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 231 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 232 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 233 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 234 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 235 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 236 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 237 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 238 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 239 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 240 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 241 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 242 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 243 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 244 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 245 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 246 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 247 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 248 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 249 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 250 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 251 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 252 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 253 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 254 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 255 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 256 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 257 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 258 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 259 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 260 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 261 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 262 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 263 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 264 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 265 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 266 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 267 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 268 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 269 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 270 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 271 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 272 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 273 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 274 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 275 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 276 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 277 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 278 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 279 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 280 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 281 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 282 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 283 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 284 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 285 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 286 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 287 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 288 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 289 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 290 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 291 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 292 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 293 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 294 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 295 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 296 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 297 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 298 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 299 *************
centroid assumed at coords: 31.0 35.0
0.0 bpix in total, and 0.0 corrected.
************Frame # 300 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 301 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 302 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 303 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 304 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 305 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 306 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 307 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 308 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 309 *************
centroid assumed at coords: 31.0 35.0
0.0 bpix in total, and 0.0 corrected.
************Frame # 310 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 311 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 312 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 313 *************
centroid assumed at coords: 31.0 35.0
8.0 bpix in total, and 8.0 corrected.
************Frame # 314 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 315 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 316 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 317 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 318 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 319 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 320 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 321 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 322 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 323 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 324 *************
centroid assumed at coords: 31.0 35.0
8.0 bpix in total, and 8.0 corrected.
************Frame # 325 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 326 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 327 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 328 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 329 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 330 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 331 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 332 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 333 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 334 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 335 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 336 *************
centroid assumed at coords: 31.0 35.0
10.0 bpix in total, and 10.0 corrected.
************Frame # 337 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 338 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 339 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 340 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 341 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 342 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 343 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 344 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 345 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 346 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 347 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 348 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 349 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 350 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 351 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 352 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 353 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 354 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 355 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 356 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 357 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 358 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 359 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 360 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 361 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 362 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 363 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 364 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 365 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 366 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 367 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 368 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 369 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 370 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 371 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 372 *************
centroid assumed at coords: 31.0 35.0
2.0 bpix in total, and 2.0 corrected.
************Frame # 373 *************
centroid assumed at coords: 31.0 35.0
1.0 bpix in total, and 1.0 corrected.
************Frame # 374 *************
centroid assumed at coords: 31.0 35.0
8.0 bpix in total, and 8.0 corrected.
************Frame # 375 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 376 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 377 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 378 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 379 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 380 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 381 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 382 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 383 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 384 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 385 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 386 *************
centroid assumed at coords: 31.0 35.0
7.0 bpix in total, and 7.0 corrected.
************Frame # 387 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 388 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 389 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 390 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 391 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 392 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
************Frame # 393 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 394 *************
centroid assumed at coords: 31.0 35.0
4.0 bpix in total, and 4.0 corrected.
************Frame # 395 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 396 *************
centroid assumed at coords: 31.0 35.0
3.0 bpix in total, and 3.0 corrected.
************Frame # 397 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 398 *************
centroid assumed at coords: 31.0 35.0
5.0 bpix in total, and 5.0 corrected.
************Frame # 399 *************
centroid assumed at coords: 31.0 35.0
6.0 bpix in total, and 6.0 corrected.
Let’s visualize the total bad pixel maps (i.e. summed over all spectral channels), and an example of concentric annuli in which bad pixels are identified.
[69]:
plot_frames((np.sum(bpm_mask,axis=0), annuli[0]))

Let’s now check one frame of the cube before and after bad pixel correction (we saturate a bit the core to better see the bad pixel):
[70]:
idx = 236
plot_frames((cube[idx], cube_corr[idx]),
vmin=float(np.amin(cube[idx])), vmax=300)

Let’s now consider a Gaussian kernel for correction of the identified bad pixels. Note that for a better correction locally, we choose the FWHM of the Gaussian kernel to be half the FWHM of the PSF:
[71]:
if version.parse(vvip) >= version.parse("1.3.1"):
cube_corr = cube_fix_badpix_interp(cube, bpm_mask, mode='gauss', fwhm=fwhm/2., half_res_y=True)
else:
cube_corr = cube_fix_badpix_with_kernel(cube, bpm_mask, mode='gauss', fwhm=fwhm/2., half_res_y=True)
Let’s check again the same frame of the cube, before and after bad pixel correction with the Gaussian kernel, and the difference between them:
[72]:
idx = 236
plot_frames((cube[idx], cube_corr[idx]),
vmin=float(np.amin(cube[idx])), vmax=300)

In this specific case, it is unclear which of the annulus-based or Gaussian kernel-based correction leads to the best correction. Keep however in mind that the best options to use for bad pixel correction will depend on your data.
2.3.4. Vertical resampling¶
Now that bad pixels are corrected, we can resample the images vertically. This can be done easily with the cube_px_resampling function. Before that, we first downsample the image vertically by a factor 2 (since every pair of rows is identical):
[73]:
nz, ny, nx = cube.shape
# downsample
nny = ny//2
cube_do = np.zeros([nz, nny, nx])
for z in range(nz):
for y in range(nny):
cube_do[z,y] = 2*cube_corr[z,y*2]
# Note: factor 2 is for flux preservation since we are only keeping every other row.
Now let’s upsample by a factor 2. It is possible to preserve the star at the center of the image with the keep_center option (i.e. the star centroid will continue to follow VIP’s convention of being at dim//2, dim//2 even when the resampling factor converts the image from even dimensions to odd dimensions). Since the star is not yet centered, we do not use that option. The scale parameter can be a float (identical factor along both x and y axes) or a tuple of two values.
[74]:
# upsample
cube_up = cube_px_resampling(cube_do, scale=(1,2), imlib='opencv', interpolation='lanczos4', keep_center=False,
verbose=True)
Cube successfully rescaled
New shape: (400, 64, 64)
Note that we used ‘opencv’ instead of the default ‘vip-fft’ for the value of imlib because FFT-based image scaling is not (yet) available in the case of different scale factors along x and y.
2.3.5. Recentering with 2D Gaussian fits¶
Now that the images have been correctly resampled vertically, we can accurately find the centroid of the star by fitting 2D Gaussians to each image:
[75]:
crop_sz = 20
nproc = None
cube_cen, shifts_y, shifts_x = cube_recenter_2dfit(cube_up, xy=(int(mean_x), int(mean_y)),
fwhm=fwhm, subi_size=crop_sz,
model='gauss', nproc=nproc,
imlib='vip-fft', interpolation=None,
full_output=True, plot=True)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2022-06-08 00:28:28
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Shifting
0% [##############################] 100% | ETA: 00:00:00
Total time elapsed: 00:00:00
Running time: 0:00:00.920806
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――

Let’s compare the images before and after resampling+recentering:
[76]:
plot_frames((cube_corr[0], cube_corr[200], cube_corr[-1],
cube_cen[0], cube_cen[200], cube_cen[-1]), rows=2)

Finally it is worth noting that this pre-processing procedure can be further refined now that the star location is known - e.g. by correcting residual bad pixels using cube_fix_badpix_ifs which requires realigned cubes and knowledge of the star location.