1B. Quick-start with object-oriented support
Authors: Thomas Bédrine and Valentin ChristiaensSuitable for VIP v1.4.2 and Python v3.10 onwardsLast update: 2024/03/04
Table of contents
This quick-start tutorial shows:
how to load Angular Differential Imaging (ADI) datacubes;
how to use algorithms implemented in VIP to model and subtract the stellar halo, and search for faint circumstellar companions in the post-processed images;
how to quickly assess the significance of a point source found in a post-processed image.
Let’s assume VIP is properly installed. If not, follow the instructions here. We import the package along with other useful packages which are installed together with VIP. The hciplot package aims to be the “Swiss army” solution for plotting and visualizing multi-dimensional high-contrast imaging datacubes.
[1]:
import vip_hci as vip
from hciplot import plot_frames, plot_cubes
from matplotlib import pyplot as plt
import numpy as np
In the following box we check that your version of VIP is recent enough to run this notebook. It should be >=1.4.2. If not, close the notebook and run for example pip install vip_hci --upgrade in your terminal. Also make sure the Python version used is 3.10 or higher.
[2]:
from packaging import version
vvip = vip.__version__
print("VIP version: ", vvip)
if version.parse(vvip) < version.parse("1.4.2"):
msg = "Please upgrade your version of VIP"
msg+= "It should be 1.4.2 or above to run this notebook."
raise ValueError(msg)
from platform import python_version
vpy = python_version()
print("Python version: ", vpy)
if version.parse(vpy) < version.parse("3.10.0"):
msg = "To use this notebook, you'll need to upgrade your Python version to 3.10 or above."
raise ValueError(msg)
VIP version: 1.6.0
Python version: 3.10.13
There are some object-oriented options to display your results, but hciplot allows lots of customization and interaction with the outputs. Interactive windows can also be generated with DS9 - more information on that in 1A - Quick start.
1.1. Loading ADI data
In order to demonstrate the capabilities of VIP, you can find in the ‘dataset’ folder of the VIP_extras repository: a toy Angular Differential Imaging (ADI) datacube of images and a point spread function (PSF) associated to the NACO instrument of the Very Large Telescope (VLT). This is an L’-band VLT/NACO dataset of beta Pictoris published in Absil et al. (2013), obtained using the Annular Groove Phase Mask (AGPM)
Vortex coronagraph. The sequence has been heavily sub-sampled temporarily to make it smaller. The frames were also cropped to the central 101x101 area. In case you want to plug-in your cube just change the path of the following cells.
Let’s first inspect the fits file:
[3]:
from vip_hci.fits import info_fits
info_fits('../datasets/naco_betapic_cube_cen.fits')
Filename: ../datasets/naco_betapic_cube_cen.fits
No. Name Ver Type Cards Dimensions Format
0 PRIMARY 1 PrimaryHDU 7 (101, 101, 61) float32
The fits file contains an ADI datacube of 61 images 101x101 px across.
The parallactic angle vector for this observation is saved in the naco_betapic_pa.fits file.
Now we load all the data in memory with the Dataset object:
[4]:
psfname = '../datasets/naco_betapic_psf.fits'
cubename = '../datasets/naco_betapic_cube_cen.fits'
angname = '../datasets/naco_betapic_pa.fits'
[5]:
from vip_hci.objects import Dataset
betapic = Dataset(cube=cubename, psf=psfname, angles=angname)
Cube array shape: (61, 101, 101)
Angles array shape: (61,)
PSF array shape: (39, 39)
Dataset uses the open_fits function, which is a wrapper of the astropy fits functionality. The fits file is now a ndarray or numpy array in memory that are stored as attributes of betapic.
If you open the datacube with ds9 (or send it to the DS9 window) and adjust the cuts you will see the beta Pic b planet moving on a circular trajectory, among similarly bright quasi-static speckle. Alternatively, you can use hciplot.plot_cubes:
[6]:
%matplotlib inline
plot_cubes(betapic.cube, vmin=0, vmax=400)
:Dataset [x,y,time] (flux)
:Cube_shape [101, 101, 61]
[6]:
The Full-Width Half-Maximum (FWHM) will automatically be computed and stored as the attribute fwhm while we do another operation : normalizing the PSF. This can be done with the function normalize_psf of the Dataset object :
[7]:
betapic.normalize_psf(size=19, imlib='ndimage-fourier')
betapic.psf = betapic.psfn
Mean FWHM: 4.801
Flux in 1xFWHM aperture: 1.307
Normalized PSF array shape: (19, 19)
The attribute `psfn` contains the normalized PSF
`fwhm` attribute set to
4.801
From now on the FWHM can simply be accessed with:
[8]:
betapic.fwhm
[8]:
4.800919383981533
Let’s visualize the normalized PSF with hciplot.plot_frames. Feel free to adapt the backend argument throughout the notebook: 'matplotlib' allows paper-quality figures with annotations which can be saved (default), while 'bokeh' enables interactive visualization.
[9]:
%matplotlib inline
plot_frames(betapic.psf, grid=True, size_factor=4) #, backend='bokeh')
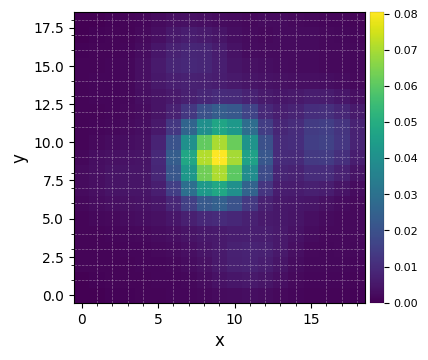
Let’s finally define the pixel scale for NACO (L’ band), which we get from a dictionary stored in the config subpackage:
[10]:
from vip_hci.config import VLT_NACO
betapic.px_scale = VLT_NACO["plsc"]
print(betapic.px_scale)
0.02719
1.2. Model PSF subtraction for ADI
When loading the ADI cube above, we assumed all calibration and preprocessing steps were already performed. In particular, the star is assumed to be already centered in the images. If your data require calibration/preprocessing, feel free to check tutorial 2. Pre-processing.
IMPORTANT - VIP’s convention regarding centering is: - for odd number of pixels along the x and y directions: the star is placed on the central pixel; - for even number of pixels: the star is placed on coordinates (dim/2,dim/2) in 0-based indexing.
1.2.1. median-ADI
The most simple approach is to model the PSF with the median of the ADI sequence, subtract it, then derotate and median-combine all residual images (Marois et al. 2006). We can do this through the PPMedianSub object, obtained with a MedianBuilder:
[11]:
from vip_hci.objects import MedianBuilder
medsub_obj = MedianBuilder(dataset=betapic).build()
medsub_obj.run()
ff_med = medsub_obj.frame_final
No changes were made to the dataset.
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2024-03-04 23:03:28
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Median psf reference subtracted
Done derotating and combining
Running time: 0:00:02.002915
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
directory element SKIPPED: _doc_link_template , calculations list : {}
directory element SKIPPED: _repr_html_ , calculations list : {}
This can also be done in concentric annuli of annular width asize, considering a parallactic angle threshold delta_rot and the nframes closest images complying with that criterion as model for subtraction to each image of the ADI cube:
[12]:
from dataclass_builder import update
update(medsub_obj, MedianBuilder(asize=betapic.fwhm, mode='annular', delta_rot=0.5, radius_int=4, nframes=4))
medsub_obj.run()
ann_med = medsub_obj.frame_final
No changes were made to the dataset.
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2024-03-04 23:03:30
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
N annuli = 9, FWHM = 4.800919383981533
Processing annuli:
0% [##############################] 100% | ETA: 00:00:00
Total time elapsed: 00:00:00
Optimized median psf reference subtracted
Done derotating and combining
Running time: 0:00:02.161983
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Let’s visualize the final images:
[13]:
plot_frames((ff_med, ann_med))
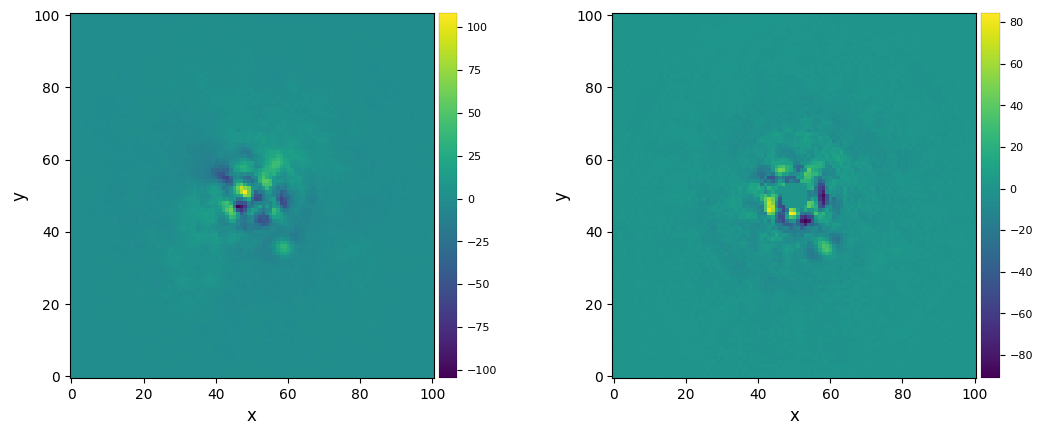
Strong residuals can be seen near the center of the image, with a faint potential point source in the south-west of the final image.
1.2.2. Full-frame PCA
Now let’s try the Principal Component Analysis (PCA) algorithm (Amara & Quanz 2012, Soummer et al. 2012), and set to 5 the number of principal components ncomp considered for model creation.
[15]:
from vip_hci.objects import PCABuilder
pca_obj = PCABuilder(dataset=betapic, ncomp=5).build()
pca_obj.run()
No changes were made to the dataset.
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2024-03-04 23:04:00
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
System total memory = 17.180 GB
System available memory = 3.444 GB
Done vectorizing the frames. Matrix shape: (61, 10201)
Done SVD/PCA with numpy SVD (LAPACK)
Running time: 0:00:00.102353
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Done de-rotating and combining
Running time: 0:00:02.211952
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
directory element SKIPPED: _doc_link_template , calculations list : {}
directory element SKIPPED: _repr_html_ , calculations list : {}
directory element SKIPPED: _doc_link_template , calculations list : {}
directory element SKIPPED: _repr_html_ , calculations list : {}
These attributes were just calculated:
directory element SKIPPED: _doc_link_template , calculations list : {}
directory element SKIPPED: _repr_html_ , calculations list : {}
frame_final
cube_reconstructed
cube_residuals
cube_residuals_der
pcs
cube_residuals_per_channel
cube_residuals_per_channel_der
cube_residuals_resc
final_residuals_cube
medians
dataframe
opt_number_pc
directory element SKIPPED: _doc_link_template , calculations list : {}
directory element SKIPPED: _repr_html_ , calculations list : {}
The following attributes can be calculated now:
detection_map with .make_snrmap()
Let’s visualize the final image. This time let’s set the grid option on, to better read coordinates from the image:
[16]:
plot_frames(pca_obj.frame_final, grid=True)
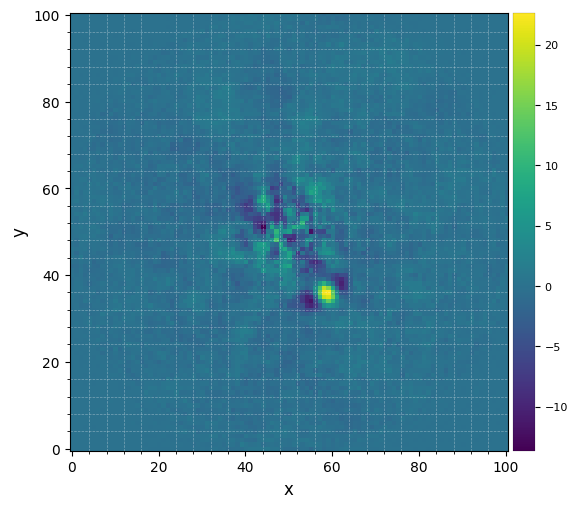
The improvement is clear compared to previous reductions. Very low residuals are seen near the star, and the companion candidate appears as a bright point source surrounded by negative side-lobes.
1.3. Is it significant?
Now let’s see whether the candidate is significant. For that let’s first set the x,y coordinates of the test point source based on the above image:
[17]:
xy_b = (58.5,35.5)
Now with this point and the previously obtained frame, we are able to compute the significance of that candidate, by using the built-in function compute_significance. This function needs at least the coordinates of the candidate and a corrected frame. If no detection map was generated through make_snrmap, it will be created inside compute_significance.
[18]:
pca_obj.compute_significance(source_xy=xy_b)
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
Starting time: 2024-03-04 23:04:09
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
S/N map created using 5 processes
Running time: 0:00:02.416477
――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――――
At a separation of 16.8 px (3.5 FWHM), S/N = 7.7 corresponds to a 5.2-sigma detection in terms of Gaussian false alarm probability.
5.2 sigma detection
Congratulations! The detection is significant! You obtained a conspicuous direct image of an exoplanet! Although from this image alone (at a single wavelength) one cannot disentangle a true physically bound companion from a background star, this source has now been extensively studied: it corresponds to the giant planet Beta Pic b (e.g. Lagrange et al. 2009, Absil et al. 2013).
You’re now ready for more advanced tutorials. If you wish to know more about:
pre-processing functionalities available in VIP (star centering, bad frame trimming, bad pixel correction…) –> go to Tutorial 2;
more advanced post-processing algorithms –> go to Tutorial 3A or 3B (the latter for Object-Oriented example usage);
metrics to evaluate the significance of a detection or the achieved contrast in your image –> go to Tutorial 4;
how to characterize a directly imaged companion –> go to Tutorial 5A;
how to characterize a circumstellar disk –> go to Tutorial 5B;
how to characterize a companion in a disk using JWST observations –> go to Tutorial 5C (to come soon);
post-processing algorithms leveraging reference star differential imaging –> go to Tutorial 6 (to come soon);
post-processing algorithms leveraging spectral differential imaging –> go to Tutorial 7;
the difference between FFT- and interpolation-based image operations (rotation, shifts, scaling) –> go to Tutorial 8;